Site-Specific Data Item (SSDI) Manual
Effective with Cases Diagnosed 1/1/2018 and Forward
Published October 2022
Version 3.0
Editors: Jennifer Ruhl, MSHCA, RHIT, CCS, CTR, NCI SEER
Jim Hofferkamp, CTR, NAACCR
Suggested Citation: Ruhl J, Hofferkamp J, et al. (October 2022). Site-Specific Data Item (SSDI)
Manual. NAACCR, Springfield, IL 62704-4194
Funding for this project was made possible in part by a contract with Federal funds from the National
Cancer Institute, National Institutes of Health and Department of Health & Human Services under
Contract number HHSN261201400004I / HHSN26100002. Additionally, funding for this project was
made possible in part by a cooperative agreement with Federal funds from the Centers for Disease
Control and Prevention Cooperative Agreement number 5NU58DP004917. Its contents are solely the
responsibility of the authors and do not necessarily represent the official views of the NCI and CDC. The
NAACCR Board of Directors adopted these standards in February 2018.

2 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
NAACCR gratefully acknowledges the dedicated work of the 2020-2022 NAACCR Site-Specific Data Item
(SSDI) Work Group.
Jennifer Ruhl, MSHCA, RHIT, CCS, CTR (NCI SEER) (chair)
Melissa Alvarado (NPCR)
Mary Brant, BS, CTR (California Cancer Registry)
Sheila Fukumura, CTR (Manitoba Cancer Registry)
Daisy Gray (Kentucky Cancer Registry)
Donna Gress, RHIT, CTR (AJCC)
Donna M. Hansen, CTR (California Cancer Registry)
Jim Hofferkamp, CTR (NAACCR)
Mei-Chin Hsieh, PhD (Louisiana Cancer Registry)
Annette Hurlbut, RHIT, CTR (Elekta)
Suzanne Kessler, MSM, RHIT, CTR (American College of Surgeons)
Richard Moldwin, M.D., Ph.D (College of American Pathologists)
Serban Negoita, MD, DrPH, CPH, CTR (NCI SEER)
Lisa Pareti, BS, RHIT, CTR (Louisiana Cancer Registry)
Loria Pollack, MD, MPH (Centers for Disease Control and Prevention)
Nicola Schussler, BS (IMS)
Aleisha Williams, MBA, CTR (AJCC)
Janine Smith, BS, CTR (California Cancer Registry)
Special Acknowledgements
Carolyn Callaghan, CTR and Tiffany Janes, CTR from the SEER*Educate program, and to Denise Harrison,
CTR (Trainer, Educatory) for their continued contribution to the SSDI Work Group
The AJCC Expert Panels for their continued critical support in clarifying concepts from the AJCC Cancer
Staging Manual, Eighth Edition and AJCC Cancer Staging System Version 9.
The College of American Pathologists (CAP) for their continued support with a CAP representative on the
SSDI Work Group and the recent participation by the CAP Cancer Committee to deal with specific issues.
CAP participation allows us to harmonize data elements between AJCC, NAACCR and the CAP Cancer
Protocols (CCPs), and electronic Cancer Checklists (eCCs). Since the terminology on many pathology
reports is guided by the latest CPPs and eCCs, the new CAP-consistent language in many of the SSDI
value sets and notes will ease the burden of coding current pathology terminology into exact matches
with NAACCR value sets. This is part of a broader effort to work towards improving interoperability
between EHR data sets and NAACCR SSDIs.
The following individuals contributed to document support and web development.
Suzanne Adams, BS, CTR (IMS)
Kathy Conklin, MSCS, Manger of IT, AJCC
Dustin Dennison, M.MIS (Information Technology Administrator, NAACCR)
Chuck May, BS (IMS)
Daniel Oluwadare, Programmer, AJCC
Nicola Schussler, BS (IMS)
3 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Table of Contents
Organization of the SSDI Manual and Suggestions for How to Use it ........................................................ 13
Introduction ................................................................................................................................................ 15
Timing for collection of SSDIs ..................................................................................................................... 17
Consult Reports ........................................................................................................................................... 18
General Definitions and Format of SSDI Codes........................................................................................... 19
Source Documents ...................................................................................................................................... 20
General Rules for Entering Laboratory Values and Other Measurements ................................................. 21
Rules for Recording Laboratory Values ....................................................................................................... 24
Rules for Recording Tests Based on Solid Tissue ........................................................................................ 28
Histologic Examination................................................................................................................................ 30
Schema Discriminators ............................................................................................................................... 31
3926: Schema Discriminator 1 ....................................................................................................... 32
3927: Schema Discriminator 2 ....................................................................................................... 33
3928: Schema Discriminator 3 ....................................................................................................... 34
SSDIs Required for Stage ............................................................................................................................. 35
SSDIs used for EOD Derived Stage Group ................................................................................................... 36
3800: Schema ID (2018+) ............................................................................................................................ 37
Schema ID Table ........................................................................................................................... 38
HEAD AND NECK ......................................................................................................................................... 48
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+) .......... 49
3926: Schema Discriminator 1: Occult Head and Neck Lymph Nodes ............................................... 49
3831: Extranodal Extension Head and Neck Clinical ........................................................................... 53
3832: Extranodal Extension Head and Neck Pathological .................................................................. 56
3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other) ........................................... 59
3876: LN Head and Neck Levels I-III .................................................................................................... 61
3877: LN Head and Neck Levels IV-V .................................................................................................. 63
3878: LN Head and Neck Levels VI-VII ................................................................................................ 65
3879: LN Head and Neck Other........................................................................................................... 67
3883: LN Size ....................................................................................................................................... 69
00071: Lip (2018+) ........................................................................................................................ 71
00072: Tongue Anterior (2018+) ................................................................................................... 71
00073: Gum (2018+) ..................................................................................................................... 71
00074: Floor of Mouth (2018+) ..................................................................................................... 71
4 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00075: Palate Hard (2018+)........................................................................................................... 71
00076: Buccal Mucosa (2018+) ...................................................................................................... 72
00077: Mouth Other (2018+) ........................................................................................................ 72
00080: Major Salivary Glands (2018+) ........................................................................................... 72
00090: Nasopharynx (2018+) ........................................................................................................ 72
3926: Schema Discriminator 1: Nasopharynx/Pharyngeal Tonsil ....................................................... 73
00100: Oropharynx HPV-Mediated (p16+) (2018+) ........................................................................ 74
3927: Schema Discriminator 2: Oropharyngeal p16 ........................................................................... 75
00111: Oropharynx (p16-) (2018+) ................................................................................................ 76
00112: Hypopharynx (2018+) ........................................................................................................ 77
00121: Maxillary Sinus (2018+) ..................................................................................................... 77
00122: Nasal Cavity and Ethmoid Sinuses (2018+) ......................................................................... 77
00130: Larynx Other (2018+) ......................................................................................................... 77
00131: Larynx Supraglottic (2018+) ............................................................................................... 77
00132: Larynx Glottic (2018+) ....................................................................................................... 77
00133: Larynx SubGlottic (2018+) .................................................................................................. 78
00140: Mucosal Melanoma of the Head and Neck (2018+) ............................................................. 78
00150: Cutaneous Carcinoma of the Head and Neck (2018+) ......................................................... 78
3858: High Risk Histologic Features .................................................................................................... 79
GASTROINTESTINAL TRACT (UPPER AND LOWER) ...................................................................................... 81
00161: Esophagus (including GE junction) Squamous (2018+) ........................................................ 82
3926: Schema Discriminator 1: EsophagusGEJunction (EGJ)/Stomach .............................................. 82
3927: Schema Discriminator 2: Histology Discriminator for 8020/3 .................................................. 85
3829: Esophagus and EGJ Tumor Epicenter ........................................................................................ 86
3855: HER2 Overall Summary ............................................................................................................. 88
00169: Esophagus (including GE junction) (excluding Squamous) (2018+) ....................................... 90
00170: Stomach (2018+) ............................................................................................................... 90
00190: Appendix (2018-2022) ....................................................................................................... 90
09190: Appendix (2023+) .............................................................................................................. 90
3960: Histologic Subtype .................................................................................................................... 91
00200: Colon and Rectum (2018+)................................................................................................. 93
3819, 3820: CEA Pretreatment Lab Value and Interpretation ............................................................ 93
3820: CEA Pretreatment Lab Value..................................................................................................... 94
3819: CEA Pretreatment Interpretation ............................................................................................. 96
5 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3823: Circumferential Resection Margin (CRM) ................................................................................. 98
3866: KRAS ........................................................................................................................................ 101
3890: Microsatellite Instability (MSI)................................................................................................104
3909: Perineural Invasion ................................................................................................................. 107
3934: Tumor Deposits ....................................................................................................................... 109
3940: BRAF Mutational Analysis ....................................................................................................... 111
3941: NRAS Mutational Analysis ....................................................................................................... 113
09210: Anus (2023+) ................................................................................................................... 116
3956: p16 .......................................................................................................................................... 116
HEPATOBILIARY SYSTEM ........................................................................................................................... 117
00220: Liver (2018+) ................................................................................................................... 118
3809, 3810: Alpha-Fetoprotein (AFP) Pretreatment Lab Value and Interpretation (Liver) .............. 118
3810: AFP Pretreatment Lab Value ................................................................................................... 119
3809: AFP Pretreatment Interpretation ........................................................................................... 120
3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score ............................ 121
3813: Bilirubin Pretreatment Total Lab Value .................................................................................. 124
3814: Bilirubin Pretreatment Unit of Measure ................................................................................. 126
3824: Creatinine Pretreatment Lab Value ........................................................................................ 127
3825: Creatinine Pretreatment Unit of Measure .............................................................................. 129
3860: International Normalized Ratio .............................................................................................. 130
3835: Fibrosis Score .......................................................................................................................... 131
002300: Bile Ducts Intrahepat (2018+) ........................................................................................ 133
3917: Primary Sclerosing Cholangitis ................................................................................................ 134
3935: Tumor Growth Pattern............................................................................................................ 136
00242: Cystic Duct (2018+) .......................................................................................................... 138
3926: Schema Discriminator 1: BileDuctsDistal/BileDuctsPerihilar/CysticDuct ............................... 138
00250: Bile Ducts Perhilar (2018+) .............................................................................................. 140
00260: Bile Duct Distal (2018+) ................................................................................................... 140
00280: Pancreas (2018+) ............................................................................................................. 141
3942: CA 19-9 PreTx Lab Value ......................................................................................................... 141
00290: NET Stomach (2018+) ...................................................................................................... 143
3863: Ki-67 ........................................................................................................................................ 143
00301: NET Duodenum (2018+)................................................................................................... 145
00302: NET Ampulla of Vater (2018+) .......................................................................................... 145
00310: NET Jejunum and Ileum (2018+)....................................................................................... 145
6 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00320: NET Appendix (2018+) ..................................................................................................... 145
00330: NET Colon and Rectum (2018+) ........................................................................................ 145
00340: NET Pancreas (2018+) ...................................................................................................... 145
THORAX ..................................................................................................................................................... 146
00360: Lung (2018+) ................................................................................................................... 147
3929: Separate Tumor Nodules ........................................................................................................ 147
3937: Visceral and Parietal Pleural Invasion ..................................................................................... 150
3938: ALK Rearrangement ................................................................................................................ 153
3939: EGFR Mutational Analysis ....................................................................................................... 155
00370: Pleura Mesothelioma (2018+) .......................................................................................... 157
3913: Pleural Effusion ....................................................................................................................... 157
BONE ......................................................................................................................................................... 159
00381: Bone Appendicular Skeleton (2018+) ............................................................................... 160
3908: Percent Necrosis Post Neoadjuvant........................................................................................ 160
00382: Bone Spine (2018+) ......................................................................................................... 162
00383: Bone Pelvis (2018+) ......................................................................................................... 162
SOFT TISSUE SARCOMA ............................................................................................................................ 163
00400: Soft Tissue Head and Neck (2018+) .................................................................................. 164
3815: Bone Invasion .......................................................................................................................... 164
00410: Soft Tissue Trunk and Extremities (2018+) ........................................................................ 166
3927: Schema Discriminator 2: Soft Tissue Sarcoma (C473, C475, C493-C495) ............................... 167
00421: Soft Tissue Abdomen and Thoracic (2018+) ...................................................................... 171
00422: Heart, Mediastinum and Pleura (2018+)........................................................................... 171
00430: GIST (2018+) ................................................................................................................... 172
3926: Schema Discriminator 1: Primary Peritoneum Tumor ............................................................ 172
3865: KIT Gene Immunohistochemistry ........................................................................................... 173
00440: Retroperitoneum (2018+) ................................................................................................ 175
00458: Soft Tissue Rare (2018+) .................................................................................................. 175
00459: Soft Tissue Other (2018+) ................................................................................................ 175
SKIN ........................................................................................................................................................... 176
00460: Merkel Cell Carcinoma (2018+) ........................................................................................ 177
3830: Extranodal Extension Clin (non-Head and Neck) .................................................................... 177
3833: Extranodal Extension Path (non-Head and Neck) ................................................................... 179
3880: LN Isolated Tumor Cells (ITC) .................................................................................................. 181
7 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3918: Profound Immune Suppression .............................................................................................. 183
00470: Melanoma Skin (2018+) ................................................................................................... 185
3817: Breslow Tumor Thickness ....................................................................................................... 185
3936: Ulceration ................................................................................................................................ 188
3893: Mitotic Rate Melanoma .......................................................................................................... 190
3932: LDH Lab Value ......................................................................................................................... 192
3869: LDH Level ................................................................................................................................. 194
3870: LDH Upper Limits of Normal ................................................................................................... 195
3961: Clinical Margin Width .............................................................................................................. 197
BREAST ...................................................................................................................................................... 199
00480: Breast (2018+) ................................................................................................................. 200
Estrogen Receptor and Progesterone Receptor ............................................................................... 200
3827: Estrogen Receptor Summary .................................................................................................. 202
3826: Estrogen Receptor Percent Positive or Range ........................................................................ 204
3828: Estrogen Receptor Total Allred Score ..................................................................................... 206
3915: Progesterone Receptor Summary ........................................................................................... 208
3914: Progesterone Receptor Percent Positive or Range ................................................................. 210
3916: Progesterone Receptor Total Allred Score ............................................................................. 212
HER2 .................................................................................................................................................. 214
3850: HER2 IHC Summary ................................................................................................................. 217
3854: HER2 ISH Summary ................................................................................................................. 219
3855: HER2 Overall Summary ........................................................................................................... 221
3853: HER2 ISH Single Probe Copy Number ..................................................................................... 223
3851: HER2 ISH Dual Probe Copy Number........................................................................................ 225
3852: HER2 ISH Dual Probe Ratio ..................................................................................................... 227
Multigene Signature Method and Results ........................................................................................ 229
3894: Multigene Signature Method .................................................................................................. 231
3895: Multigene Signature Results ................................................................................................... 233
Oncotype Dx Tests ............................................................................................................................ 235
3903: Oncotype Dx Recurrence Score-DCIS ...................................................................................... 237
3905: Oncotype Dx Risk Level-DCIS .................................................................................................. 238
3904: Oncotype Dx Recurrence Score-Invasive ................................................................................ 239
3906: Oncotype Dx Risk Level-Invasive ............................................................................................. 241
3863: Ki-67 ........................................................................................................................................ 242
3882: LN Positive Axillary Level I-II ................................................................................................... 244
8 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3922: Response to Neoadjuvant Therapy ......................................................................................... 247
FEMALE REPRODUCTIVE ORGANS ............................................................................................................ 249
3836: FIGO ........................................................................................................................................ 250
Lymph Node Assessment Methods & Status for Regional & Distant Lymph Nodes in GYN Sites .... 253
00500: Vulva (2018+) .................................................................................................................. 254
3836: FIGO: Vulva ............................................................................................................................. 254
3959: LN Status: Femoral-Inguinal .................................................................................................... 255
3871: LN Assessment Method Femoral-Inguinal .............................................................................. 257
3957: LN Status: Pelvic ...................................................................................................................... 259
3873: LN Assessment Method Pelvic ................................................................................................ 261
3881: LN Laterality ............................................................................................................................ 263
00510: Vagina (2018+) ................................................................................................................ 265
3836: FIGO: Vagina............................................................................................................................ 265
3959: LN Status: Femoral-Inguinal .................................................................................................... 266
3871: LN Assessment Method Femoral-Inguinal .............................................................................. 268
3958: LN Status: Para-Aortic ............................................................................................................. 270
3872: LN Assessment Method Para-Aortic ....................................................................................... 272
3957: LN Status: Pelvic ...................................................................................................................... 274
3873: LN Assessment Method Pelvic ................................................................................................ 276
3875: LN Distant: Mediastinal, Scalene ............................................................................................ 278
3874: LN Distant Assessment Method .............................................................................................. 279
00520: Cervix (2018-2020) .......................................................................................................... 280
3836: FIGO: Cervix............................................................................................................................. 281
09520: Cervix (2021+) ................................................................................................................. 283
3956: p16 .......................................................................................................................................... 284
00528: Cervix Sarcoma ............................................................................................................... 285
00530: Corpus Carcinoma and Carcinosarcoma (2018+) ............................................................... 286
3836: FIGO: Corpus Carcinoma and Carcinosarcoma ....................................................................... 286
3899-3902: Number of Positive and Examined Para-Aortic and Pelvic Nodes ................................. 288
3901: Number of Positive Para-Aortic Nodes ................................................................................... 289
3899: Number of Examined Para-Aortic Nodes ................................................................................ 291
3902: Number of Positive Pelvic Nodes ............................................................................................ 293
3900: Number of Examined Pelvic Nodes ......................................................................................... 295
3911: Peritoneal Cytology ................................................................................................................. 297
00541: Corpus Sarcoma (2018+) .................................................................................................. 299
9 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3836: FIGO Stage (Sarcoma) ............................................................................................................. 300
00542: Corpus Adenosarcoma (2018+) ........................................................................................ 301
3836: FIGO Stage (Adenosarcoma) ................................................................................................... 302
00551: Ovary (2018+) ................................................................................................................. 303
3836: FIGO: Ovary, Fallopian Tube, and Peritoneal Carcinoma ........................................................ 303
3818: CA-125 Pretreatment Interpretation ...................................................................................... 305
3921: Residual Tumor Volume Post Cytoreduction .......................................................................... 307
00552: Primary Peritoneal Carcinoma (2018+) ............................................................................. 309
00553: Fallopian Tube (2018+) .................................................................................................... 310
00560: Placenta (2018+) ............................................................................................................. 311
3836: FIGO: Gestational Trophoblastic Tumors (Placenta) .............................................................. 311
3837: Gestational Trophoblastic Prognostic Scoring Index .............................................................. 312
MALE GENITAL ORGANS ........................................................................................................................... 314
00570: Penis (2018+) .................................................................................................................. 315
00580: Prostate (2018+) ............................................................................................................. 316
3920: PSA (Prostatic Specific Antigen) Lab Value ............................................................................. 316
Gleason Patterns and Scores ............................................................................................................ 319
3838: Gleason Patterns Clinical ........................................................................................................ 322
3840: Gleason Score Clinical ............................................................................................................. 325
3839: Gleason Patterns Pathological ................................................................................................ 327
3841: Gleason Score Pathological ..................................................................................................... 330
3842: Gleason Tertiary Pattern ......................................................................................................... 332
Number of Cores Positive and Examined .......................................................................................... 333
3898: Number of Cores Positive ....................................................................................................... 334
3897: Number of Cores Examined .................................................................................................... 336
00590: Testis (2018+) .................................................................................................................. 338
Testis Serum Markers and S Category .............................................................................................. 338
Alpha-fetoprotein (AFP) (Testis) ....................................................................................................... 339
3807: AFP Pre-Orchiectomy Lab Value ............................................................................................. 341
3808: AFP Pre-Orchiectomy Range ................................................................................................... 343
3805: AFP Post-Orchiectomy Lab Value ............................................................................................ 345
3806: AFP Post-Orchiectomy Range ................................................................................................. 347
Human Chorionic Gonadotropin (hCG) (Testis) ................................................................................ 349
3848: hCG Pre-Orchiectomy Lab Value ............................................................................................. 351
3849: hCG Pre-Orchiectomy Range .................................................................................................. 352
10 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3846: hCG Post-Orchiectomy Lab Value ........................................................................................... 353
3847: hCG Post-Orchiectomy Range ................................................................................................. 355
Lactate Dehydrogenase (LDH) (Testis)..............................................................................................357
3868: LDH Pre-Orchiectomy Range................................................................................................... 359
3867: LDH Post-Orchiectomy Range ................................................................................................. 360
3923: S Category Clinical ................................................................................................................... 362
3924: S Category Pathological .......................................................................................................... 364
URINARY TRACT ........................................................................................................................................ 366
00600: Kidney Parenchyma (2018+) ............................................................................................ 367
3864: Invasion Beyond Capsule ........................................................................................................ 367
3886: Major Vein Involvement ......................................................................................................... 369
3861: Ipsilateral Adrenal Gland Involvement ................................................................................... 371
3925: Sarcomatoid Features ............................................................................................................. 373
00631: Urethra (2018+) .............................................................................................................. 375
3926: Schema Discriminator 1: Urethra/Prostatic Urethra .............................................................. 375
00633: Urethra-Prostatic (2018+) ................................................................................................ 376
OPHTHALMIC SITES ................................................................................................................................... 377
00640: Skin Eyelid (2018+) .......................................................................................................... 378
3909: Perineural Invasion ................................................................................................................. 378
00660: Melanoma Conjunctiva (2018+) ....................................................................................... 380
00671: Melanoma Iris (2018+) .................................................................................................... 381
3926: Schema Discriminator 1: Melanoma Ciliary Body/Melanoma Iris .......................................... 381
3821: Chromosome 3 Status ............................................................................................................. 382
3822: Chromosome 8q Status ........................................................................................................... 384
3834: Extravascular Matrix Patterns ................................................................................................. 386
3887: Measured Basal Diameter ....................................................................................................... 388
3888: Measured Thickness ............................................................................................................... 390
3891: Microvascular Density ............................................................................................................. 392
3892: Mitotic Count Uveal Melanoma .............................................................................................. 394
00672: Melanoma Choiroid and Ciliary Body (2018+) ................................................................... 396
00680: Retinoblastoma (2018+) .................................................................................................. 397
3856: Heritable Trait ......................................................................................................................... 397
00690: Lacrimal Gland (2018+) .................................................................................................... 399
3926: Schema Discriminator 1: Lacrimal Gland/Sac ......................................................................... 400
3803: Adenoid Cystic Basaloid Pattern ............................................................................................. 401
11 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00698: Lacrimal Sac (2018+) ........................................................................................................ 403
CENTRAL NERVOUS SYSTEM ..................................................................................................................... 404
00721: Brain (2018-2022) ............................................................................................................ 405
3816: Brain Molecular Markers ........................................................................................................ 405
3801, 3802: Loss of Heterozygosity: Chromosome 1p and Chromosome 19q (CNS) ....................... 407
3801: Chromosome 1p: Loss of Heterozygosity (LOH) ..................................................................... 408
3802: Chromosome 19q: Loss of Heterozygosity (LOH) ................................................................... 410
3889: Methylation of O6-Methylguanine-Methyltransferase .......................................................... 412
09721: Brain (2023+) .................................................................................................................. 414
00722: CNS Other (2018-2022) .................................................................................................... 415
09722: CNS Other (2023+) ........................................................................................................... 416
09724: Medulloblastoma (2023+) ............................................................................................... 417
ENDOCRINE SYSTEM ................................................................................................................................. 418
00730: Thyroid (2018+) ............................................................................................................... 419
3926: Schema Discriminator 1: Thyroid Gland/Thyroglossal Duct ................................................... 419
00740: Thyroid Medullary (2018+) .............................................................................................. 420
HEMATOLOGIC MALIGNANCIES ................................................................................................................ 421
00790: Lymphoma (excluding CLL/SLL) (2018+) ........................................................................... 422
3926: Schema Discriminator 1: Histology Discriminator for 9591/3 ................................................ 422
3812: B Symptoms ............................................................................................................................ 423
3859: HIV Status ................................................................................................................................ 425
3896: NCCN International Prognostic Index (IPI) .............................................................................. 427
00795: Lymphoma-CLL/SLL (2018+) ............................................................................................. 429
Rai Classification ............................................................................................................................... 430
3885: Lymphocytosis......................................................................................................................... 432
3804: Adenopathy ............................................................................................................................. 434
3907: Organomegaly ......................................................................................................................... 436
3811: Anemia .................................................................................................................................... 438
3933: Thrombocytopenia .................................................................................................................. 440
3955: Derived Rai stage .................................................................................................................... 442
00811: Mycosis Fungoides (2018+) .............................................................................................. 443
3910: Peripheral Blood Involvement ................................................................................................ 443
00821: Plasma Cell Myeloma (2018+) .......................................................................................... 446
RISS Stage (Plasma Cell Myeloma) .................................................................................................... 446
3926: Schema Discriminator 1: Plasma Cell Myeloma Terminology ................................................ 447
12 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3931: Serum Beta-2 Microglobulin Pretreatment Level ................................................................... 448
3930: Serum Albumin Pretreatment Level ....................................................................................... 449
3857: High Risk Cytogenetics............................................................................................................450
3869: LDH Level ................................................................................................................................. 451
00830: HemeRetic (2018+) .......................................................................................................... 452
3862: JAK 2 ........................................................................................................................................ 453
99999: Ill-Defined Other (2018+) ................................................................................................. 455
ALPHABETICAL INDEX ............................................................................................................................... 456

13 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Organization of the SSDI Manual and Suggestions for How to Use it
The Site-Specific Data Item (SSDI) manual is the primary resource for documentation and coding
instructions for site-specific data items introduced in 2018. Information in the SSDI Manual is similar to
that provided in the Collaborative Stage v2 (CSv2) Manual Part I, Section II for Site Specific Factors (SSF).
Before using the Manual as an information resource for specific data items, it is important to review the
introductory materials and general instructions carefully. Although the majority of data items that are
collected as SSDIs were previously collected as SSFs, the format of the data items and allowable values
have changed substantially, particularly for laboratory values.
Information about the SSDIs has been organized using primary site groupings and presented in the order
used in the AJCC Manuals, a format that is familiar and useful to registrars and most others using the
SSDI Manual. However, we have also provided an alphabetical index for the SSDIs with the
corresponding page number in the last 2 pages of the Manual for those who may want to search it for a
specific SSDI by data item name. The Table of Contents for the Manual contains hyperlinks so that
clicking anywhere on the line where an SSDI and page number are listed will take you directly to that
page in the Manual.
An important new concept introduced in 2018 is the use of a Schema ID to define the applicable SSDIs
and grade table for a particular tumor, based on primary site, histology, and in some cases, additional
information. The appropriate Schema ID will be defined by registry software and will not have to be
assigned by the registrar. However, a Schema ID Table defining the Schema ID number, description and
associated SSDIs is provided in the SSDI Manual for reference purposes. The Schema ID Table will also be
useful for registrars abstracting cases before their software is available. In addition to Schema IDs, the
Schema ID Table also provides the applicable SSDIs with a hyperlink to the page on which the
description of the relevant SSDIs begins. A hyperlink at the end of the information on each SSDI can be
used to return to the Schema ID Table.
For each SSDI, the SSDI Manual includes:
NAACCR Data Item Name
Item Length
NAACCR Item #
NAACCR Alternative Name
Schema ID(s)
Description
o The description is a brief summary used to define the data item in the NAACCR data
dictionary
Rationale
o The rationale describes the reason why the data item is collected, such as required for
staging or recommended for registry data collection by AJCC. If the data item was
collected in CSv2, the primary site and SSF# is included in the rationale

14 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Definition
o The definition provides additional background on the data item and its clinical
importance. This information was previously included in the CSv2 Manual, Part I, Section
II
Additional Information
o This section may include source documents, other names, normal reference ranges and
any other information deemed relevant for a particular SSDI. This information was
previously included in the CSv2 Manual, Part I, Section II
Coding instructions and Codes
o Coding instructions are provided as numbered notes. Codes are provided in a table.
Codes and coding instructions are usually provided in registry software.
Appendix A
Appendix A, presented in Schema ID order, provides detailed information on the sites, histologies and
behavior codes included in each schema, along with the applicable SSDIs, grade table, EOD Schema
Name, Summary Stage 2018 Chapter and the current AJCC Staging System. This information is used in
registry software development and may also be useful to researchers and others interested in
understanding schema definitions.
Appendix B
Appendix B is an excel spreadsheet which lists all of the CSv2 site specific factors by CS Schema, their
current status (based on CoC), primary site, and (where applicable), the NAACCR v18 Data Item # and
Name.
Appendix C
Appendix C is a WORD document which lists all the SSDIs in numerical order, the applicable Schema ID(s)
and the start and end year.

15 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Introduction
In 2018, Collaborative Stage (CS) Site-Specific Factors (SSFs) will be discontinued and Site-Specific Data
Items (SSDIs) will be used for collection of site-specific information. SSDIs will have unique names and
NAACCR data item numbers and can be applied to as many sites as needed. Unlike SSFs, field length is
not limited to 3 digits, decimals are allowed, and different coding conventions are used to record actual
values, percentages and ranges. NAACCR is the custodian of the SSDIs and the SSDI TF is responsible for
their development and updates.
The NAACCR Site-Specific Data Item Taskforce
In October 2016, the NAACCR Site-Specific Data Item (SSDI) Taskforce was formed to determine how to
collect information recorded in the site-specific factors (SSFs) which are part of the Collaborative Stage
software (CS DLL). The taskforce evaluated the structure of the CS SSFs and made recommendations on
how the information should be collected and then updated/revised the format, codes, and coding
instructions as needed.
Taskforce members evaluated several different ways of collecting the information. The final decision
was to discontinue the CS SSF approach and create new individual site-specific data items (SSDIs) for
data collection beginning with cases diagnosed in 2018. There are several reasons for this decision.
More flexibility
No longer will all site-specific data items be three characters in length. Some are shorter, others
are longer
Also, registrars can record lab values with the decimal point as part of the code.
Meaningful names
Each new data item has been given a name that will be displayed in registry software.
o For example, the software displays ER instead of Breast, SSF1
It is easier for registrars and researchers to retrieve data.
o For example, query the database for PSA instead of remembering that SSF1 is PSA in
Prostate
Reduced duplication
CS SSFs which were collected for multiple sites/chapters/schema under different SSF numbers
are now one data item when possible
What is a SSDI?
A “SSDI’” is a site-specific data item. “Site” in this instance is based on the primary site, the AJCC Staging
System, Summary Stage chapter and the EOD schema. SSDIs were preceded by CS SSFs, which were first
introduced in 2004 with CSv1, and went through major revisions in 2010 with Collaborative Stage v2
(CSv2). CS SSFs were discontinued as of 12/31/2017.
SSDIs have their own data item name and number and can be collected for as many sites/systems
/schemas as needed.
Each Site-Specific Data Item (SSDI) applies only to selected schemas. SSDI fields should be blank for
schemas where they do not apply.
Unless otherwise noted, all SSDIs start collection in 2018. For those that have a collection start date later
than 2018, a note has been added to instruct registrars when it should be collected.

16 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
How SSDIs are associated with relevant site/histologies and schemas
In Collaborative Stage v2 (CSv2), 153 Schemas were defined based on site/histology and used to assign
applicable site-specific factors (SSFs) and staging algorithms. For 2018, Schema ID [3800] is used to link
all combinations of sites and histologies (using additional information from schema discriminators if
needed) with the appropriate stage data collection systems and SSDIs. AJCC ID [995] is used to link AJCC
staging eligible sites/histologies (using additional information from schema discriminators if needed)
with the appropriate AJCC Staging System and staging algorithm. Schema ID and AJCC ID will be derived
by registry software based on site and histology codes entered by the registrar. Refer to Appendix A for
a complete listing of schemas IDs and related schema information.
Process of Developing the SSDIs
Development of the SSDIs began with reviewing the CS SSFs. Due to the number of CS SSFs, and the fact
that many of them were discontinued in CSv0204, a priority order was established.
First: schema discriminators. These are data items needed to determine the correct SSDIs, AJCC
Staging System, EOD schema, or Summary Stage schema
Second: data items required to assign stage
Third: data items currently required by at least one standard setter and listed as registry
collection data items in at least one AJCC Staging System
Last: certain data items required by standard setters and not necessarily stage related. These
comprise a small percentage of the data items
CS SSFs discontinued in CSv0204 were not reviewed for 2018 data collection. New registry data
collection items listed in the AJCC Staging System was not reviewed, unless they are required for staging.
Number of SSDIs compared to CS SSFs
Approximately 260 unique CS SSFs in CSv0205
101 discontinued
12 obsolete
147 required
Of these, 27 are not required for 1/1/2018+
120 SSDIs added to the NAACCR v18 layout
CS SSF data will be retained for cases diagnosed 2004-2017. CS SSF data will not be mapped to the SSDIs.
Collection of CS SSFs or the new SSDIs is based strictly on the date of diagnosis. For cases
diagnosed 2004-2017, CS SSFs will continue to be collected according to the appropriate
standard setter. For cases diagnosed 2018 or later, the SSDIs will be collected according to the
appropriate standard setter
Example: A case diagnosed in 2017 is abstracted in 2018. Code the applicable/required CS SSFs for
that case, not the SSDIs.
For a complete listing of site-specific factors from CSv0205 and the corresponding SSDI (if any) for 2018,
see Appendix B.
17 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Timing for collection of SSDIs
The SSDIs are to be collected during the initial diagnosis, work up and first course of treatment. Some
SSDIs have specific instructions as to when the SSDIs are collected (e.g., CEA is to be collected prior to
polypectomy, or PSA is to be collected prior to needle core biopsy).
Note: Active surveillance is first course of treatment.

18 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Consult Reports
If a report is sent out for consult and the results are different than the original report, record the results
from the consult
Example 1: Patient had biopsy done at a facility with a Gleason Score of 4+4=8. Slides were sent out for
consult and their review showed Gleason Score 4+3=7.
Record the Gleason score of 4+3=7 based on the consult.
Example 2: Original pathology report states ER and PR positive. Slides were sent out for consult and their
review showed ER and PR negative.
Record ER and PR as negative
Example 3: Breast pathology report states Grade 3, ER 95% strong on outside pathology. Patient
presents at facility for treatment and the slides from the outside facility are reviewed, with the results of
Grade 2, ER 80% intermediate.
Record Grade 2 and ER 80% intermediate

19 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
General Definitions and Format of SSDI Codes
Not applicable: This code is to be used ONLY when the data item is relevant for the case and the
standard setter does not require the data item. Not applicable codes ALWAYS end in an 8 but will differ
depending on the length of the data item.
Note: “Not applicable” is not available for schema discriminators or data items which are required
for staging.
Examples:
Perineural Invasion. This is a 1-digit field. “Not applicable” is 8
FIGO Stage (for all GYN cases). This is a 2-digit field. “Not applicable” is 98
Creatinine Pretreatment Lab Value. This is a 4-digit field including the decimal point. “Not
applicable” is XX.8
AFP (Alpha Fetoprotein) Pre-Orchiectomy Lab Value. This is a 7-digit field including the decimal
point. “Not applicable” is XXXXX.8
It is important to review each data item carefully to determine how the “not applicable” code is
formatted.
Unknown: Previous codes from CS for test not done (998) and unknown (999) have been combined.
Unknown codes ALWAYS end in a 9 but will differ depending on the length of the data item. The
unknown code includes
Test/evaluation/assessment not done or UNKNOWN if done
“Cannot be determined by pathologist.” For some data items, this is a selection box on the College of
American Pathologists (CAP) checklist. Cannot be determined by pathologist is primarily used when a
tissue specimen is not adequate for testing.
“Not identified.” For some data items, this is a selection box on the CAP checklist. This means that the
pathologist has looked for it and it is not present. This is not the same thing as looking for it in the
medical record and not finding it (this would be “not documented in the medical record.”)
Death Certificate Only (DCOs) cases
For DCOs, the applicable SSDIs (except for applicable Schema Discriminators) may be blank.
Note: This instruction is for central registries only.

20 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Source Documents
Source documents are suggested for some data items as the most likely sources of information.
If no source document is suggested, use any information provided in the medical record
If a pathology report is suggested, that document includes
o Addenda or revisions to the report
o Gross or microscopic description
o Synoptic reports
o CAP protocol, or cancer checklist information provided by the pathologist
It is important to review each data item carefully to determine where the information can be found. For
some data items, the information is based on imaging or some other type of clinical exam. Other data
items are based on pathological findings from a surgical resection.

21 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
General Rules for Entering Laboratory Values and Other Measurements
Lab values and other measurements that are not integers (whole numbers) and are reported as
continuous variables (not categories or ranges) will be recorded to a single decimal place with an explicit
decimal point.
There must always be a numeral or the letter ‘X’ immediately before the decimal point and a numeral
after the decimal point, which will be in the next-to-last character position in the field. The entered
value must be right-justified in the field and padded with spaces to the left if necessary to fill the field.
Users’ software will usually justify and pad the value automatically for the registrar.
In addition to the actual values, codes are defined for situations such as value unknown; test done but
results not in chart; and other special cases. Sometimes codes will be provided for when a value is
expressed as “at least” some value.
These may be needed, for example, in the measurement of tumor size or thickness when the
tumor has been transected and the actual size cannot be determined. These codes will begin
with one or more ‘X’s.
When a value in the medical record does not provide the expected decimal digit, i.e. it is expressed as a
whole number, then enter the value followed by a decimal point and a zero.
Examples for a 6-Character Lab Value
Value in Record Data Item Coded as
0.0 0.0
0 0.0
.1 0.1
11.0 11.0
11.1 11.1
11 11.0
111.1 111.1
1111.1 1111.1

22 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Rounding Rules
SSDIs follow the standard definitions for rounding. These general rules can be followed for most SSDIs
where lab values or percentages are recorded. All SSDIs that have lab values, percentages or
measurements are set up to record in the 10ths (one digit after the decimal point). If a lab value,
percentage or measurement is recorded in 100ths (two digits after the decimal point), then the last digit
must be rounded.
The general rounding rules are:
If digit is 0-4, round down
If digit is 5-9, round up
Note: Currently (2018+), the only SSDIs that have exceptions to the general rounding rules are:
o HER2 ISH Single Probe Copy Number
o HER2 ISH Dual Probe Copy Number
o HER2 ISH Dual Probe Ratio
Examples
Breslow’s measurement 4.32 mm
o Since the last digit is 2, round down and record 4.3
CEA lab value 18.35
o Since the last digit is 5, round up and record 18.4
HER2 ISH Dual Probe Copy Number 6.78
o Per note 8: If the test results are presented to the hundredth decimal, ignore the
hundredth decimal. Do NOT round. Record 6.7
o This also applies to HER2 ISH Single Probe Copy Number and HER2 ISH Dual Probe Ratio
Note: ER (and PR) percent positive do not have decimal points in the data items, so anything
with a decimal point will have to be rounded.
o Example: 78.6. Since the last digit is 6, round up and record 079 (79%)
o Note: For ER and PR percent positive, if a value is documented as 99.5% to 99.9%, round
up to 100% (code 100)
23 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Recording values when “less than” or “greater than” are used
Record the value as one less than stated when a value is reported as “less than X,” and as one more
than stated when a value is reported as “more than X.” One less or one more may refer to a whole
number (1), or a decimal (0.1), depending on the code structure of the field.
SSDIs with decimals in their code structures
Example 1: PSA stated as < (less than) 5. Record 4.9
Example 2: hCG lab value resulting findings of < (less than) 1. Record 0.9
Example 3: Ki-67 reported as > (greater than) 20%. Record 20.1
SSDIs without decimals in their code structure:
Example 1: ER Percent Positive stated as < (less than) 60%. Record 059 (59%)
Example 2: PR Percent Positive stated as > (greater than) 75%. Record 076 (76%)
Example 3: ER Percent Positive < (less than) 50%. Record 049 (49%)

24 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Rules for Recording Laboratory Values
Laboratory values refer to any tests that are based on blood, urine, ascites, or spinal fluid. Most of these
are based on blood.
Do not apply these rules to SSDIs that are based on tissue; see Rules for Recording Tests Based on Solid
Tissue .
Follow the below guidelines for recording laboratory values:
All laboratory values must be done no earlier than approximately three months before diagnosis
Only record test results obtained before any cancer-directed treatment is given (neoadjuvant
therapy or surgical), unless instructions for a specific laboratory test state otherwise
Record the highest laboratory value if multiple laboratory tests results are available, unless
instructions for a specific laboratory test state otherwise
The following SSDIs record laboratory values. If the SSDI specific coding rules column is yes, then check
the SSDI for additional coding instructions
Schema SSDI# SSDI SSDI Specific Coding
Rules
Colon and Rectum 3820 CEA Pretreatment Lab Value Yes
Colon and Rectum 3819 CEA Pretreatment Interpretation Yes
Liver 3810 AFP Pretreatment Lab Value
Liver 3809 AFP Pretreatment Interpretation
Liver 3813 Bilirubin Pretreatment Total Lab Value
Liver 3814 Bilirubin Pretreatment Unit of Measure
Liver 3820 Creatinine Pretreatment Total Lab Value
Liver 3825 Creatinine Pretreatment Unit of Measure
Liver 3860 International Normalized Ratio for Prothrombin Time
Lymphoma (CLL/SLL) 3811 Anemia
Lymphoma (CLL/SLL) 3933 Thrombocytopenia
Mycosis Fungoides 3910 Peripheral Blood Involvement
Ovary, Fallopian Tube,
Primary Peritoneal
Carcinoma
3818 CA-125 Pretreatment Interpretation
Melanoma Skin 3932 LDH Lab Value Yes
Melanoma Skin 3869 LDH Level Yes
Melanoma Skin 3870 LDH Upper Limits of Normal Yes
Pancreas 3942 CA 19-9 PreTx Lab Value
Plasma Cell Myeloma 3930 Serum Albumin Pretreatment Level
Plasma Cell Myeloma 3931 Serum Beta-2 Microglobulin Pretreatment Level
Plasma Cell Myeloma 3932 LDH Lab Value
Prostate 3920 PSA Lab Value (See SSDI specific instructions) Yes
Testis 3807 AFP Pre-Orchiectomy Lab Value Yes
Testis 3808 AFP Pre-Orchiectomy Range Yes
Testis 3805 AFP Post-Orchiectomy Lab Value Yes
Testis 3806 AFP Post-Orchiectomy Range Yes
Testis 3848 hCG Pre-Orchiectomy Lab Value Yes
Testis 3849 hCG Pre-Orchiectomy Range Yes
Testis 3846 hCG Post-Orchiectomy Lab Value Yes
Testis 3847 hCG Post-Orchiectomy Range Yes

25 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema SSDI# SSDI SSDI Specific Coding
Rules
Testis 3868 LDH Pre-Orchiectomy Range Yes
Testis 3867 LDH Post-Orchiectomy Range Yes
If the only test or tests performed do not meet these criteria, code "test not done" or "unknown if test
performed."
The results of laboratory values vary according to the laboratory conducting the test. The normal
reference range is included in the tumor marker comments as background information only. Some data
items ask for a laboratory value, others ask for the “interpretation” of the laboratory test (normal,
elevated, and so forth).
When the data item asks for the interpretation of a laboratory test, code the clinician’s/pathologist’s
interpretation, if available, as first priority. This would include statements of “abnormal”, “elevated”,
“normal”, “equivocal”, “present”, “absent”, and so forth. In addition, the physician's statement of a T,
N, or M value or stage group for the case could be an implied interpretation of a lab value used to
determine the TNM classification.
Example 1: Physician summarizes breast cancer workup by saying "HER2 IHC was positive at
3+.” Registrar would code interpretation as positive
Note: If the pathologist uses the term "indeterminate," code as borderline; undetermined if
positive or negative if that code exists in the data item. If a code for borderline or
undetermined does not exist, code as unknown
In the absence of a physician’s interpretation of the test, if the reference range for the lab is listed on
the test report, the registrar may use that information to assign the appropriate code.
Example 2: Medical record laboratory report shows ovarian cancer patient's CA-125 as 69
(normal range < 35 U/ml). Registrar may infer that CA-125 is elevated
When there is no clinician/pathologist interpretation of the lab test and no description of the reference
range in the medical record the registrar should code unknown. Do not code the lab value
interpretation based on background information provided in this manual for the data item.
Note: There will be some cases where an interpretation may be inferred from the background
information in this manual because the lab result is extremely abnormal. In such cases, common
sense would dictate that the case should be coded as elevated rather than unknown.
Example 3: Physician reports that Alpha Fetoprotein (AFP) collected in the office for a patient
suspected to have primary liver cancer was 750 but does not interpret this value. Background
information in the manual indicates a high normal would be > 500 but hepatocellular carcinoma
values are > 1000. Registrar should code AFP Interpretation as unknown
Example 4: Physician reports a CEA of 450 for a colon cancer without interpreting
it. Background information in the manual indicates a high normal would be 5 ng/ml. Registrar
may code CEA as elevated

26 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
What does SI mean? SI is the French abbreviation for International System (Systeme Internationale),
standard units of measure (meter, kilogram, second). Most SI values are based on the kilogram and the
liter. A nanogram (ng) is one-thousandth of a microgram . A milliliter (ml) is one-thousandth of a
liter. Therefore, a lab value expressed in mg/L is equivalent to the same value expressed in
ng/ml. Some lab values, such as hormone levels, are recorded in International Units per Liter
(IU/L). This is equivalent to mIU/mL. The equivalence of mIU to ng varies according to what is
measured.
Note that instructions for entering many lab values state that the registrars should not convert the
values. For those where conversion is allowed, one measurement conversion website is:
https://www.amamanualofstyle.com/page/si-conversion-calculator
SI Conversion: 1 mg/L = 1 ng/ml.
For example, 1 ng of AFP is approximately equal to 1 mIU.
Note: Micrograms per liter may be printed as ug/L.
Prefixes and abbreviations. Units of measure can be described and written in various ways in the
medical record. In some circumstances, the unit of measure may be dependent on the printer used for
the report.
For example, the prefix “micron” (one millionth of a unit) is represented in scientific notation by
the Greek letter mu (m), but not all printers have the capability to print Greek symbols. As a
result, micro- may be printed as a lower-case u or as the abbreviation mc.
Do not confuse the abbreviation for micro- (u) with the abbreviation for Unit (an international
system measurement, U).

27 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Tables I-2-1a – I-2-1c below show abbreviations for units of measurement and the abbreviations for
fractions or multiples of those units.
Table I-2-1a. Measurement Prefixes
Number Prefix Written
1,000,000 Mega- M
1000 Kilo- k
10
Deka
-
da
1 (baseline)
1/10 Deci- d
1/100 Centi- c
1/1000 Milli- m
One millionth Micro- m, u, or mc
One billionth Nano- n
One trillionth
Pico
-
p
One quadrillionth Femto f
Table I-2-1b. Unit Abbreviations
Unit Abbrev.
Liter L
Unit U
Meter
m
Unit-of-substance mole, mol
Gram g, gr
milli-Equivalent mEq, meq
Table I-2-1c. Examples
Unit Abbrev.
Femtomole
fmol
Microgram ugr, mcg, mgr
Milliliter ml

28 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Rules for Recording Tests Based on Solid Tissue
Priority Order for SSDIs
Addendums or amendments (corrections that are not incorporated into the initial synoptic
report, including CAP Cancer Protocol)
Synoptic report (including CAP Cancer Protocol)
Pathology report: final diagnosis
Physician statement
General Rules versus SSDI specific rules
Unless instructions for a specific tissue test state otherwise, record the highest value (positive
versus negative, or actual numerical value) obtained from any tissue based examination (biopsy,
surgical resection, bone marrow biopsy).
If the SSDI specific coding rules column is yes, then check the SSDI for additional coding
instructions
Schema SSDI# SSDI SSDI Specific
Coding Rules
Bile Ducts Intrahepatic 3935 Tumor Growth Pattern
Bile Ducts Intrahepatic
Liver
3835 Fibrosis Score
Bile Ducts Intrahepatic,
Bile Ducts Perihilar
3917 Primary Sclerosing Cholangitis
Bone 3908 Percent Necrosis Post Neoadjuvant Yes
Brain, CNS 3816 Brain Molecular Markers
Brain, CNS 3801 Chromosome 1p: Loss of Heterozygosity
Brain, CNS 3802 Chromosome 19q: Loss of Heterozygosity
Brain, CNS 3889 Methylation of O6-Methylguanine-Methyltransferase
(MGMT)
Breast 3827 Estrogen Receptor Summary Yes
Breast 3826 Estrogen Receptor Percent Positive or Range Yes
Breast 3828 Estrogen Receptor Total Allred Score Yes
Breast 3882 LN Positive Axillary Level I-II Yes
Breast 3915 Progesterone Receptor Summary Yes
Breast 3914 Progesterone Receptor Percent Positive or Range Yes
Breast 3916 Progesterone Receptor Total Allred Score Yes
Breast 3855 HER2 Overall Summary Yes
Breast 3894 Multigene Signature Method
Breast 3895 Multigene Signature Results
Breast 3903 Oncotype Dx Recurrence Score-DCIS
Breast 3905 Oncotype Dx Risk Level-DCIS
Breast 3904 Oncotype Dx Recurrence Score-Invasive
Breast 3906 Oncotype Dx Risk Level Invasive
Breast 3863 Ki-67 Yes
Colon and Rectum 3823 Circumferential Resection Margin Yes
Colon and Rectum 3866 KRAS
Colon and Rectum 3890 Microsatellite Instability (MSI)
Colon and Rectum 3909 Perineural Invasion Yes
Colon and Rectum 3934 Tumor Deposits Yes
Colon and Rectum 3940 BRAF Mutational Analysis
Colon and Rectum 3941 NRAS Mutational Analysis

29 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema SSDI# SSDI SSDI Specific
Coding Rules
Cutaneous Carcinoma
Skin
3858 High Risk Histologic Features
Cutaneous Carcinoma
Skin
3909 Perineural Invasion Yes
Esophagus (both
schemas)
3855 HER2 Overall Summary
GIST 3865 KIT Gene Immunochemistry
HemeRetic 3862 JAK2
Kidney 3864 Invasion Beyond Capsule Yes
Kidney 3886 Major Vein Involvement Yes
Kidney 3861 Ipsilateral Adrenal Gland Involvement Yes
Kidney 3925 Sarcomatoid Features
Lacrimal Gland 3803 Adenoid Cystic Basaloid Pattern
Lacrimal Gland 3903 Perineural Invasion Yes
Lung 3938 ALK Rearrangement
Lung 3939 EGFR Mutational Analysis
Lung 3937 Visceral and Parietal Pleural Invasion
Melanoma Choroid and
Ciliary Body; Iris
3821 Chromosome 3 status
Melanoma Choroid and
Ciliary Body; Iris
3821 Chromosome 8q status
Melanoma Choroid and
Ciliary Body; Iris
3834 Extravascular Matrix Patterns
Melanoma Choroid and
Ciliary Body; Iris
3887 Measured Basal Diameter
Melanoma Choroid and
Ciliary Body; Iris
3888 Measured Thickness
Melanoma Choroid and
Ciliary Body; Iris
3891 Microvascular Density
Melanoma Choroid and
Ciliary Body; Iris
3892 Mitotic Count Uveal Melanoma
Melanoma Skin 3817 Breslow Tumor Thickness Yes
Melanoma Skin 3936 Ulceration Yes
Melanoma Skin 3893 Mitotic Rate Melanoma
NET Schemas 3863 Ki-67 Yes
Plasma Cell Myeloma 3857 High Risk Cytogenetics
Prostate 3838 Gleason Patterns Clinical Yes
Prostate 3840 Gleason Score Clinical Yes
Prostate 3839 Gleason Patterns Pathological Yes
Prostate 3841 Gleason Score Pathological Yes
Prostate 3898 Number of Cores Positive Yes
Prostate 3897 Number of Cores Examined
Yes
Retinoblastoma 3856 Heritable Trait
Stomach 3855 HER2 Overall Summary
30 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Histologic Examination
Histologic examination is the assessment of a tissue specimen. Aspiration of fluid (cells) is a cytologic
examination. Some data items require analysis of tissue, whereas others can be performed on any
specimen (tissue or fluid). Pathological examination can refer to either histological or cytological
examination.
Also referred to as “microscopic confirmation.”
31 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema Discriminators
Introduced in Collaborative Stage version 2 (CSv2), schema discriminators are used when primary site
and/or histology are not sufficient to identify the correct AJCC staging algorithm. Due to the complexity
of some of the AJCC Staging System, more than one schema discriminator may be needed to define the
correct schema. Three SSDIs (Data Item #’s 3926, 3927 and 3928) are available to collect the information
needed to define schema, although most sytems that require a schema discriminator need only one.
Schema discriminators are used to define both Schema ID, used to link all combinations of sites and
histologies, with the appropriate stage data collection systems and SSDIs, and AJCC ID, used to link AJCC
staging eligible sites/histologies with the appropriate AJCC Staging System and staging algorithm.
Schema discriminators do not have a “not applicable” code. If the schema discriminator is needed for
some sites or histologies within the schema but not for all, it should be left blank where it is not
necessary.

32 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3926: Schema Discriminator 1
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Description
Captures additional information needed to generate AJCC ID and Schema ID for some anatomic sites.
Discriminators can be based on sub site, histology or other features which affect prognosis.
Rationale
A schema discriminator is used to assign AJCC ID when site and histology alone are insufficient to
identify the applicable AJCC staging method and to assign Schema ID, which links each case to the
appropriate SSDIs, Summary Stage and EOD data collection system.
Codes (The information recorded in Schema Discriminator differs for each anatomic site. See the SSDI
manual for most current version of the site-specific codes and coding structures.)
The following are Schema Discriminator 1
3926: Schema Discriminator 1: BileDuctsDistal/BileDuctsPerihilar/CysticDuct
3926: Schema Discriminator 1: EsophagusGEJunction (EGJ)/Stomach
3926: Schema Discriminator 1: Histology Discriminator for 9591/3
3926: Schema Discriminator 1: Lacrimal Gland/Sac
3926: Schema Discriminator 1: Melanoma Ciliary Body/Melanoma Iris
3926: Schema Discriminator 1: Nasopharynx/Pharyngeal Tonsil
3926: Schema Discriminator 1: Occult Head and Neck Lymph Nodes
3926: Schema Discriminator 1: Plasma Cell Myeloma Terminology
3926: Schema Discriminator 1: Primary Peritoneum Tumor
3926: Schema Discriminator 1: Thyroid Gland/Thyroglossal Duct
3926: Schema Discriminator 1: Urethra/Prostatic Urethra

33 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3927: Schema Discriminator 2
Item Length: 1
NAACCR Item #: 3927
XML Parent-NAACCR ID: Tumor-schemaDiscriminator2
NAACCR Alternate Name: None
Active years: 2018+
Description
Captures additional information needed to generate AJCC ID and Schema ID for some anatomic sites.
Discriminators can be based on sub site, histology or other features which affect prognosis.
Rationale
A schema discriminator is used to assign AJCC ID when site and histology alone are insufficient to
identify the applicable AJCC staging method and to assign Schema ID, which links each case to the
appropriate SSDIs, Summary Stage and EOD data collection system.
Codes (The information recorded in Schema Discriminator differs for each anatomic site. See the SSDI
manual for most current version of the site-specific codes and coding structures.)
The following are Schema Discriminator 2
3927: Schema Discriminator 2: Histology Discriminator for 8020/3
3927: Schema Discriminator 2: Oropharyngeal p16
3927: Schema Discriminator 2: Soft Tissue Sarcoma (C473, C475, C493-C495)

34 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3928: Schema Discriminator 3
Item Length: 1
NAACCR Item #: 3928
XML Parent-NAACCR ID: Tumor-schemaDiscriminator3
NAACCR Alternate Name: None
Active years: 2018+
Description
Captures additional information needed to generate AJCC ID and Schema ID for some anatomic sites.
Discriminators can be based on sub site, histology or other features which affect prognosis.
Rationale
A schema discriminator is used to assign AJCC ID when site and histology alone are insufficient to
identify the applicable AJCC staging method and to assign Schema ID, which links each case to the
appropriate SSDIs, Summary Stage and EOD data collection system.
There are currently no defined Schema Discriminators 3s.

35 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
SSDIs Required for Stage
In addition to AJCC T, N, M or EOD fields (primary tumor, regional nodes, and mets), there are SSDIs that
are needed to either assign an AJCC stage or derive the EOD Derived Stage Group.
Note: Required for stage data items do not have a “not applicable” code. These data items must
be coded for all applicable cases. If the information is not available, code the appropriate
“unknown” value.
For further information on these data items, see the individual data items.
SSDI
#/Description
Schema ID
#/Description
3829: Esophagus and EGJ Tumor Epicenter
00161:
Esophagus (including GE junction) Squamous
3827: Estrogen Receptor Summary 00480: Breast
3915: Progesterone Receptor Summary 00480: Breast
3855: HER2 Overall Summary 00480: Breast
3904: Oncotype Dx Recurrence Score-Invasive 00480: Breast
3837: Gestational Trophoblastic Prognostic Scoring
Index
00560: Placenta
3920: PSA (Prostatic Specific Antigen) Lab Value 00580: Prostate
3923: S Category Clinical 00590: Testis
3924: S Category Pathological 00590: Testis
3856: Heritable Trait 00680: Retinoblastoma
3804: Adenopathy 00795: Lymphoma (CLL/SLL)
3811: Anemia 00795: Lymphoma (CLL/SLL)
3885: Lymphocytosis 00795: Lymphoma (CLL/SLL)
3907: Organomegaly 00795: Lymphoma (CLL/SLL)
3933: Thrombocytopenia 00795: Lymphoma (CLL/SLL)
3910: Peripheral Blood Involvement 00811: Mycosis Fungoides
3857: High Risk Cytogenetics
00821:
Plasma Cell Myeloma
3869: LDH Level
00821:
Plasma Cell Myeloma
3930: Serum Albumin Pretreatment Level 00821: Plasma Cell Myeloma
3931: Serum Beta-2 Microglobulin Pretreatment Level 00821: Plasma Cell Myeloma

36 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
SSDIs used for EOD Derived Stage Group
In addition to the SSDIs required for AJCC stage, the following SSDIs are used for the EOD Derived Stage
group. These SSDIs are only required for those registries that are collecting EOD but may be collected by
others.
SSDI#/Description Schema ID#/Description
3883: LN Size 00100: Oropharynx HPV-Mediated (p16+)
3869: LDH Level 00470: Melanoma Skin
3882: LN Positive Axillary Level I-II 00480: Breast
3911: Peritoneal Cytology 00530: Corpus Carcinoma and Carcinosarcoma
3911: Peritoneal Cytology 00541: Corpus Sarcoma
3911: Peritoneal Cytology 00542: Corpus Adenosarcoma
3887: Measured Basal Diameter 00671: Melanoma Iris
3887: Measured Basal Diameter 00672: Melanoma Choroid and Ciliary Body
3888: Measured Thickness
00671: Melanoma Iris
3888: Measured Thickness
00672: Melanoma Choroid and Ciliary
Body

37 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3800: Schema ID (2018+)
Item Length: 5
NAACCR Item #: 3800
XML Parent-NAACCR ID: Tumor-schemaID
NAACCR Alternate Name: None
Active years: 2018+
Description
The derived values in this data item link Site-Specific Data Items (including grade data items) with the
appropriate site/histology grouping and account for every combination of primary site and histology.
The values for this data item are derived based on primary site, histology, and schema discriminator
fields (when required). The derived values link Site-Specific Data Items with the appropriate
site/histology grouping.
For example, the Schema ID for an adenocarcinoma of the lung is 00360. This value links the
Site-Specific Data Items associated with adenocarcinoma of the lung: Separate Tumor Nodules
[3929], Visceral and Parietal Pleural Invasion [3937], and Pleural Effusion [3913].
The Schema ID would also link to the appropriate grade data items an adenocarcinoma of the lung. The
AJCC ID [995] code for Lung is 36. The AJCC ID [995] would link to the AJCC TNM Data items (Clin T, Clin
N, Etc.) specific to Lung. AJCC ID [995] will not be assigned when a site/histology combination is not
eligible for TNM staging.
Rationale
The purpose of the derived Schema ID is to link the appropriate Site-Specific Data Items with the
patient’s primary site/histology. This data item is similar to AJCC ID [995] but includes additional
site/histologies that may not be eligible for TNM staging using the current AJCC Staging Manual. AJCC ID
[995] is left blank if a case is not eligible for TNM Staging using the current AJCC Staging Manual.
Separating AJCC ID [995] and the Schema ID allows coding of Site-Specific Data Items for site/histology
combinations that are not eligible for an AJCC Stage but are eligible for Summary Stage. This data item
will also be used to develop edits and could potentially be used for analysis. Codes: See the NAACCR
Site-Specific Data Item webpage for codes. Each Site-Specific Data Item (SSDI) applies only to selected
primary sites, histologies, and years of diagnosis.
Definition
In Collaborative Stage v2 (CSv2), 153 Schemas were defined based on site/histology and used to assign
applicable site-specific factors (SSFs) and staging algorithms. Beginning on January 1, 2018, SSFs are
replaced with SSDIs and site-specific grading systems are used and Schema ID [3800] is used to link all
combinations of sites and histologies (using additional information from schema discriminators if
needed) with the appropriate SSDIs and site-specific grading system. A separate data item, AJCC ID
[995], is used to link AJCC staging eligible sites/histologies (using additional information from schema
discriminators if needed) with the appropriate AJCC Staging System and staging algorithm. Schema ID
and AJCC ID will be derived by registry software based on site and histology codes entered by the
registrar.

38 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID Table
Schema ID#/Description
SSDI
#/Description
Years Applicable
00060: Cervical Lymph Nodes
and Unknown Primary Tumors
of the Head and Neck (2018+)
3926: Schema Discriminator 1: Occult Head and Neck
Lymph Nodes (primary site C760)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
3883: LN Size
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00071: Lip (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00072: Tongue Anterior
(2018+)
3831
: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00073: Gum (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00074: Floor of Mouth (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00075: Palate Hard (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00076: Buccal Mucosa (2018+)
3831
: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00077: Mouth Other (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00080: Major Salivary Glands
(2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00090: Nasopharynx (2018+) 3926: Schema Discriminator 1:
Nasopharynx/PharyngealTonsil
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
2018+

39 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00100: Oropharynx HPV-
Mediated (p16+)
3926: Schema Discriminator 1:
Nasopharynx/PharyngealTonsil
3927: Schema Discriminator 2: Oropharyngeal p16
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
2018+
2018+
00111: Oropharynx (p16-)
(2018+)
3926: Schema Discriminator 1:
Nasopharynx/PharyngealTonsil
3927: Schema Discriminator 2: Oropharyngeal p16
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
2018+
2018+
00112: Hypopharynx (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00118: Pharynx Other (2018+) No SSDIs defined for this Schema ID NA
00119: Middle Ear (2018+) No SSDIs defined for this Schema ID NA
00121: Maxillary Sinus (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00122: Nasal Cavity and
Ethmoid Sinuses (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00128: Sinus Other (2018+)
No SSDIs defined for this Schema ID
NA
00130: Larynx Other (2018+)
3831
: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00131: Larynx Supraglottic
(2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00132: Larynx Glottic (2018+) 3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+
00133: Larynx SubGlottic
(2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3883: LN Size
2018+
2018+
2018+

40 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00140: Mucosal Melanoma of
the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck
Pathological
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
3883: LN Size
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00150: Cutaneous Carcinoma
of the Head and Neck (2018+)
3858
: High Risk Histologic Features
3883: LN Size
3909: Perineural Invasion
2018+
2018+
2018+
00161: Esophagus (including
GE junction) Squamous
(2018+)
3926: Schema Discriminator 1: EsophagusGE Junction
(EGJ)/Stomach) (primary site C160)
3927: Schema Discriminator 2: Histology
Discriminator for 8020/3
3829: Esophagus and EGJ Tumor Epicenter
3855: HER2 Overall Summary
2018+
2018+
2018+
2021+
00169: Esophagus (including
GE junction) (excluding
Squamous) (2018+)
3926: Schema Discriminator 1: EsophagusGE Junction
(EGJ)/Stomach) (primary site C160)
3927: Schema Discriminator 2: Histology
Discriminator for 8020/3
3855: HER2 Overall Summary
2018+
2018+
2021+
00170: Stomach (2018+) 3926: Schema Discriminator 1: EsophagusGE Junction
(EGJ)/Stomach) (primary site C160)
3855: HER2 Overall Summary
2018+
2021+
00180: Small
Intestine (2018+)
No SSDIs defined for this Schema ID
NA
00190: Appendix (2018
-
2022)
3819
: CEA Pretreatment Interpretation
3820: CEA Pretreatment Lab Value
2018
-
2022
2018-2022
09190: Appendix (2023+)
3819
: CEA Pretreatment Int
erpretation
3820: CEA Pretreatment Lab Value
3960: Histology Subtype
20
23+
2023+
2023+
00200: Colon and Rectum
(2018+)
3819: CEA Pretreatment Interpretation
3820: CEA Pretreatment Lab Value
3823: Circumferential Resection Margin (CRM)
3866: KRAS
3890: Microsatellite Instability (MSI)
3909: Perineural Invasion
3934: Tumor Deposits
3940: BRAF Mutational Analysis
3941: NRAS Mutational Analysis
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2021+
2021+
00210: Anus (2018-2022) No SSDIs defined for this Schema ID NA
09210: Anus (2023+) 3956: p 16 2023+
00220: Liver (2018+) 3809: AFP Pretreatment Interpretation
3810: AFP Pretreatment Lab Value
3813: Bilirubin Pretreatment Total Lab Value
3814: Bilirubin Pretreatment Unit of Measure
3824: Creatinine Pretreatment Lab Value
3825: Creatinine Pretreatment Unit of Measure
3835: Fibrosis Score
3860: International Normalized Ratio
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+

41 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00230: Bile Ducts Intrahepatic
(2018+)
3835: Fibrosis Score
3917: Primary Sclerosing Cholangitis
3935: Tumor Growth Pattern
2018+
2018+
2018+
00241: Gallbladder No SSDIs defined for this Schema ID
NA
00242: Cystic Duct (2018+) 3926: Schema Discriminator 1:
BileDuctsDistal/BileDuctsPerihilar/ CysticDuct
2018+
00250: Bile Ducts Perhilar
(2018+)
3926: Schema Discriminator 1:
BileDuctsDistal/BileDuctsPerihilar/ CysticDuct
3917: Primary Sclerosing Cholangitis
2018+
2018+
00260: Bile Duct Distal (2018+) 3926: Schema Discriminator 1:
BileDuctsDistal/BileDuctsPerihilar/ CysticDuct
2018+
00270: Ampulla of Vater
(2018+)
No SSDIs defined for this Schema ID NA
00278: Biliary Other (2018+) No SSDIs defined for this Schema ID NA
00280: Pancreas (2018+) 3942: CA 19-9 PreTx Lab Value 2021+
00288: Digestive Other (2018+) No SSDIs defined for this Schema ID NA
00290: NET Stomach (2018+) 3863: Ki-67
2021+
00301: NET Duodenum
(2018+)
3863: Ki-67
2021+
00302: NET Ampulla of Vater
(2018+)
3863: Ki-67
2021+
00310: NET Jejunum and Ileum
(2018+)
3863: Ki-67
2021+
00320: NET Appendix (2018+) 3863: Ki-67
2021+
00330: NET Colon and Rectum
(2018+)
3863:
Ki
-
67
2021+
00340: NET Pancreas (2018+) 3863: Ki-67
2021+
00350: Thymus No SSDIs defined for this Schema ID NA
00358: Trachea No SSDIs defined for this Schema ID NA
00360: Lung (2018+) 3929: Separate Tumor Nodules
3937: Visceral and Parietal Pleural Invasion
3938: ALK Rearrangement
3939: EGFR Mutational Analysis
2018+
2018+
2021+
2021+
00370: Pleura Mesothelioma
(2018+)
3913: Pleural Effusion 2018+
00378: Respiratory Other
(2018+)
No SSDIs defined for this Schema ID NA
00381: Bone Appendicular
Skeleton (2018+)
3908: Percent Necrosis Post Neoadjuvant 2018+
00382: Bone Spine (2018+) 3908: Percent Necrosis Post Neoadjuvant 2018+
00383: Bone Pelvis (2018+) 3908: Percent Necrosis Post Neoadjuvant 2018+
00400: Soft Tissue Head and
Neck (2018+)
3815: Bone Invasion 2018+
00410: Soft Tissue Trunk and
Extremities (2018+)
3815: Bone Invasion
3927: Schema Discriminator 2: Soft Tissue Trunk and
Extremities/ Soft Tissue Abdomen and Thoracic
2018+
2018+
00421: Soft Tissue Abdomen
and Thoracic (2018+)
3815: Bone Invasion
3927: Schema Discriminator 2: Soft Tissue Trunk and
Extremities/ Soft Tissue Abdomen and Thoracic
2018+
2018+

42 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00422: Heart, Mediastinum
and Pleura (2018+)
3815: Bone Invasion 2018+
00430: GIST (2018+) 3926: Schema Discriminator 1: Primary Peritoneum
Tumor
3865: KIT Gene Immunohistochemistry
2018+
2018+
00440: Retroperitoneum
(2018+)
3815: Bone Invasion
00458: Soft Tissue Rare
(2018+)
3815: Bone Invasion
00459: Soft Tissue Other
(2018+)
3815: Bone Invasion
3926: Schema Discriminator 1: Occult Head and Neck
Lymph Nodes (primary site C760)
3927: Schema Discriminator 2: Soft Tissue Trunk and
Extremities/ Soft Tissue Abdomen and Thoracic
00458: Kaposi Sarcoma No SSDIs defined for this Schema ID NA
00460: Merkel Cell Carcinoma
(2018+)
3830: Extranodal Extension Clin (non-Head & Neck)
3833: Extranodal Extension Path (non-Head & Neck)
3880: LN Isolated Tumor Cells (ITC)
3918: Profound Immune Suppression
2018+
2018+
2018+
2018+
00470: Melanoma Skin (2018+) 3817: Breslow Tumor Thickness
3936: Ulceration
3893: Mitotic Rate Melanoma
3932: LDH Lab Value
3869: LDH Level
3870: LDH Upper Limits of Normal
3961: Clinical Margin Width
2018+
2018+
2018+
2018+
2018+
2018+
2023+
00478: Skin Other (2018+) No SSDIs defined for this Schema ID NA
00480: Breast (2018+)
3826
: Estrogen Receptor Percent Positive or Range
3827: Estrogen Receptor Summary
3828: Estrogen Receptor Total Allred Score
3914: Progesterone Receptor Percent Positive or
Range
3915: Progesterone Receptor Summary
3916: Progesterone Total Allred Score
3850: HER2 IHC Summary
3851: HER2 ISH Dual Probe Copy Number
3852: HER2 ISH Dual Probe Ratio
3853: HER2 ISH Single Probe Copy Number
3854: HER2 ISH Summary
3855: HER2 Overall Summary
3894: Multigene Signature Method
3895: Multigene Signature Results
3903: Oncotype Dx Recurrence Score-DCIS
3904: Oncotype Dx Recurrence Score-Invasive
3905: Oncotype Dx Risk Level-DCIS
3906: Oncotype Dx Risk Level-Invasive
3863: Ki-67
3882: LN Positive Axillary Level I-II
3922: Response to Neoadjuvant Therapy
2018
+
2018+
2018-2022
2018+
2018+
2018-2022
2018-2020
2018-2020
2018-2020
2018-2020
2018-2020
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+

43 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00500: Vulva (2018+) 3836: FIGO Stage (Vulva)
3959: LN Status: Femoral-Inguinal
3871: LN Assessment Method Femoral-Inguinal
3957: LN Status: Pelvic
3873: LN Assessment Method Pelvic
3881: LN Laterality
2018+
2018+
2018+
2018+
2018+
2018+
00510: Vagina (2018+) 3836: FIGO Stage (Vagina)
3959: LN Status: Femoral-Inguinal
3871: LN Assessment Method Femoral-Inguinal
3958: LN Status: Para-aortic
3872: LN Assessment Method Para-aortic
3959: LN Status: Pelvic
3873: LN Assessment Method Pelvic
3874: LN Distant Assessment Method
3875: LN Distant: Mediastinal, Scalene
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00520: Cervix (2018-2020) 3836: FIGO Stage (Cervix)
3871: LN Assessment Method Femoral-Inguinal
3872: LN Assessment Method Para-aortic
3873: LN Assessment Method Pelvic
3874: LN Distant Assessment Method
3875: LN Distant: Mediastinal, Scalene
2018-2020
2018-2020
2018-2020
2018-2020
2018-2020
2018-2020
09520: Cervix (2021+)
3836
: FIGO Stage (Cervix)
3958: LN Status Para-aortic
3872: LN Assessment Method Para-aortic
3957: LN Status Pelvic
3873: LN Assessment Method Pelvic
3874: LN Distant Assessment Method
3875: LN Distant: Mediastinal, Scalene
3956: p 16
20
21+
2021+
2021+
2021+
2021+
2021+
2021+
2021+
00528: Cervix Sarcoma 3836: FIGO Stage (Corpus Sarcoma)
3899: Number of Examined Para-Aortic Nodes
3900: Number of Examined Pelvic Nodes
3901: Number of Positive Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3911: Peritoneal Cytology
2021+
2021+
2021+
2021+
2021+
2021+
00530: Corpus Carcinoma and
Carcinosarcoma
3836: FIGO Stage (Corpus Carcinoma and
Carcinosarcoma)
3899: Number of Examined Para-Aortic Nodes
3900: Number of Examined Pelvic Nodes
3901: Number of Positive Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3911: Peritoneal Cytology
2018+
2018+
2018+
2018+
2018+
2018+
00541: Corpus Sarcoma 3836: FIGO Stage (Corpus Sarcoma)
3899: Number of Examined Para-Aortic Nodes
3900: Number of Examined Pelvic Nodes
3901: Number of Positive Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3911: Peritoneal Cytology
2018+
2018+
2018+
2018+
2018+
2018+

44 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00542: Corpus Adenosarcoma 3836: FIGO Stage (Corpus Adenosarcoma)
3899: Number of Examined Para-Aortic Nodes
3900: Number of Examined Pelvic Nodes
3901: Number of Positive Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3911: Peritoneal Cytology
2018+
2018+
2018+
2018+
2018+
2018+
00551: Ovary (2018+) 3818: CA-125 Pretreatment Interpretation
3836: FIGO Stage (Ovary, Fallopian Tube and Primary
Peritoneal Carcinomas)
3921: Residual Tumor Volume Post Cytoreduction
2018+
2018+
2018+
00552: Primary Peritoneal
Carcinoma (2018+)
3818
: CA
-
125 Pretreatment Interpretation
3836: FIGO Stage (Ovary, Fallopian Tube and Primary
Peritoneal Carcinomas)
3921: Residual Tumor Volume Post Cytoreduction
2018
+
2018+
2018+
00553: Fallopian Tube (2018+) 3818: CA-125 Pretreatment Interpretation
3836: FIGO Stage (Ovary, Fallopian Tube and Primary
Peritoneal Carcinomas)
3921: Residual Tumor Volume Post Cytoreduction
2018+
2018+
2018+
00558: Adnexa Uterine Other
(2018+)
No SSDIs defined for this Schema ID NA
00559: Genital Female Other
(2018+)
No SSDIs defined for this Schema ID NA
00560: Placenta (2018+) 3836: FIGO Stage (Gestational Trophoblastic Tumors)
3837: Gestational Trophoblastic Prognostic Scoring
Index
2018+
2018+
00570: Penis (2018+) 3830: Extranodal Extension Clin (non-Head and Neck)
3833: Extranodal Extension Path (non-Head and
Neck)
2018+
2018+
00580: Prostate (2018+) 3838: Gleason Patterns Clinical
3839: Gleason Patterns Pathological
3840: Gleason Score Clinical
3841: Gleason Score Pathological
3842: Gleason Tertiary Pattern
3897: Number of Cores Examined
3898: Number of Cores Positive
3920: PSA (Prostatic Specific Antigen) Lab Value
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00590: Testis (2018+) 3805: AFP Post-Orchiectomy Lab Value
3806: AFP Post-Orchiectomy Range
3807: AFP Pre-Orchiectomy Lab Value
3808: AFP Pre-Orchiectomy Range
3846: hCG Post-Orchiectomy Lab Value
3847: hCG Post-Orchiectomy Range
3848: hCG Pre-Orchiectomy Lab Value
3849: hCG Pre-Orchiectomy Range
3867: LDH Post-Orchiectomy Range
3868: LDH Pre-Orchiectomy Range
3923: S Category Clinical
3924: S Category Pathological
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00598: Genital Male Other
(2018+)
No SSDIs defined for this Schema ID NA

45 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00600: Kidney Parenchyma
(2018+)
3861: Ipsilateral Adrenal Gland Involvement
3864: Invasion Beyond Capsule
3886: Major Vein Involvement
3925: Sarcomatoid Features
2018+
2018+
2018+
2018+
00610: Kidney Renal Pelvis
(2018+)
No SSDIs defined for this
Schema ID
NA
00620: Bladder (2018+) No SSDIs defined for this Schema ID NA
00631: Urethra (2018+) 3926: Schema Discriminator 1 (Urethra/Prostatic
Urethra)
2018+
00633: Urethra-Prostatic
(2018+)
3926: Schema Discriminator 1 (Urethra/Prostatic
Urethra)
2018+
00638: Urinary Other (2018+) No SSDIs defined for this Schema ID NA
00640: Skin Eyelid (2018+) 3909: Perineural Invasion 2018+
00650: Conjunctiva (2018+) No SSDIs defined for this Schema ID NA
00600: Melanoma Conjunctiva 3888: Measured Thickness 2018+
00671: Melanoma Iris (2018+) 3926: Schema Discriminator 1 (Melanoma Ciliary
Body/Melanoma Iris)
3821: Chromosome 3 Status
3822: Chromosome 8q Status
3834: Extravascular Matrix Patterns
3887: Measured Basal Diameter
3888: Measured Thickness
3891: Microvascular Density
3892: Mitotic Count Uveal Melanoma
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00672: Melanoma Choiroid
and Ciliary Body (2018+)
3926: Schema Discriminator 1 (Melanoma Ciliary
Body/Melanoma Iris)
3821: Chromosome 3 Status
3822: Chromosome 8q Status
3834: Extravascular Matrix Patterns
3887: Measured Basal Diameter
3888: Measured Thickness
3891: Microvascular Density
3892: Mitotic Count Uveal Melanoma
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00680: Retinoblastoma
(2018+)
3856: Heritable Trait 2018+
00690: Lacrimal Gland (2018+) 3926: Schema Discriminator 1 (Lacrimal
Gland/Lacrimal Sac)
3803: Adenoid Cystic Basaloid Pattern
3909: Perineural Invasion
2018+
2018+
2018+
00698: Lacrimal Sac (2018+)
3926:
Schema Discriminator 1 (Lacrimal
Gland/Lacrimal Sac)
2018
+
00700: Orbital Sarcoma
(2018+)
No SSDIs defined for this Schema ID
NA
00710: Lymphoma
Ocular
Adnexa (2018+)
No SSDIs defined for this Schema ID
NA
00718: Eye Other (2018+) No SSDIs defined for this Schema ID NA

46 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00721: Brain (2018-2022) 3801: Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity
(LOH)
3816: Brain Molecular Markers
3889: Methylation of O6-Methylguanine-
Methyltransferase
2018-2022
2018-2022
2018-2022
2018-2022
09721: Brain (2023+) 3801: Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity
(LOH)
3816: Brain Molecular Markers
3889: Methylation of O6-Methylguanine-
Methyltransferase
2023+
2023+
2023+
2023+
00722: CNS Other (2018-2022) 3801: Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity
(LOH)
3816: Brain Molecular Markers
3889: Methylation of O6-Methylguanine-
Methyltransferase
2018-2022
2018-2022
2018-2022
2018-2022
09722: CNS Other (2023+)
3801
:
Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity
(LOH)
3816: Brain Molecular Markers
3889: Methylation of O6-Methylguanine-
Methyltransferase
2023+
2023+
2023+
2023+
00723: Intracranial Other
(2018-2022)
No SSDIs defined for this Schema ID
NA
09723: Intracranial Other
(2023+)
No SSDIs defined for this Schema ID
NA
09724: Medulloblastoma
(2023+)
3816
: Brain Molecular Markers
2023+
00730: Thyroid (2018+)
3926
:
Schema Discriminator 1 (Thyroid
Gland/Thyroglossal Duct)
2018
+
00740: Thyroid Medullary
(2018+)
3926
:
Schema Discriminator 1 (Thyroid
Gland/Thyroglossal Duct)
2018
+
00750: Parathyroid (2018+) No SSDIs defined for this Schema ID NA
00760: Adrenal Gland (2018+) No SSDIs defined for this Schema ID NA
00770: NET Adrenal Gland
(2018+)
No SSDIs defined for this Schema ID NA
00778: Endocrine Other
(2018+)
No SSDIs defined for this Schema ID NA
00790: Lymphoma (excluding
CLL/SLL) (2018+)
3926: Schema Discriminator 1 (Histology
Discriminator for 9591/3)
3812: B Symptoms
3859: HIV Status
3896: NCCN International Prognostic Index (IPI)
2018+
2018+
2018+
2018+

47 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Schema ID#/Description SSDI #/Description Years Applicable
00795: Lymphoma-CLL/SLL
(2018+)
3812: B Symptoms
3859: HIV Status
3896: NCCN International Prognostic Index (IPI)
3804: Adenopathy
3811: Anemia
3812: B symptoms
3859: HIV Status
3885: Lymphocytosis
3896: NCCN International Prognostic Index (IPI)
3907: Organomegaly
3933: Thrombocytopenia
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
2018+
00811: Mycosis Fungoides
(2018+)
3910
:
Peripheral Blood Involvement
2018
+
00812: Primary Cutaneous
Lymphomas (excluding
Mycosis Fungoides) (2018+)
No SSDIs defined for this Schema ID NA
00821: Plasma Cell Myeloma
(2018+)
3926: Schema Discriminator 1 (Multiple Myeloma
Terminology)
3857: High Risk Cytogenetics
3869: LDH Level
3930: Serum Albumin Pretreatment Level
3931: Serum Beta-2 Microglobulin Pretreatment
Level
2018+
2018+
2018+
2018+
2018+
00822: Plasma Cell Disorders
(2018+)
No SSDIs defined for this Schema ID
NA
00830: HemeRetic (2018+)
3926
:
Schema Discriminator 1 (Histology
Discriminator for 9591/3)
3862: JAK2
2018
+
2018+
99999: ILL-DEFINED OTHER
(2018+)
3926: Schema Discriminator 1 (Occult Head and Neck
Lymph Nodes) (Primary site C760 only)
2018+
48 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
HEAD AND NECK

49 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3926: Schema Discriminator 1: Occult Head and Neck Lymph Nodes
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00060: Cervical Lymph Nodes and Unknown Primary (2018+)
00459: Soft Tissue Other (2018+)
99999: Ill-Defined Other (2018+)
Definition
In AJCC 8
th
edition, a new chapter was introduced for situations when there are positive cervical nodes
(head and neck nodes for Levels I-VII, and other group), however, the primary tumor is not evident
(occult tumor) and the primary tumor is suspected to be from the head and neck region (primary sites
C00-C14, C30-32).
If the differential diagnosis includes non-head and neck sites, the primary site should be coded
to C809
o Example: path report states metastasis to the cervical lymph node could be from a head
and neck primary, lung primary, or gynecologic primary
If there is no indication that the cervical lymph node is from a head and neck site, then the
primary site should be coded to C809
If the tumor is found to be EBV+ or p16+. See Coding Guidelines below.
If the tumor is suspected to be from a head and neck site, or a potential head and neck site is
indicated by the physician, see Coding Guidelines below.
To develop a software algorithm that can be used to send the registrar to the right chapter/schema, this
schema discriminator was developed.
To get to this schema discriminator, the registrar will code C760 (head and neck, NOS) when there is a
suspected head and neck tumor, yet the primary site is not known, and/or the primary tumor was not
identified. The schema discriminator will then be brought up.
Note: If the physician “suspects” or “assigns” a specific head and neck subsite, the registrar is
still to assign C760 so that the correct staging information can be abstracted.
o Example: FNA of cervical lymph node metastases shows squamous cell carcinoma, p16
negative. Workup shows no evidence of primary tumor, although physician states this
may be a laryngeal primary based on “best guess”.
Even though the primary site is suspected to be larynx, primary site would still be coded to
C760. For all head and neck sites, except for oropharynx p16+ and nasopharynx EBV positive, no
evidence of primary tumor (T0) does not exist in the individual AJCC chapters or EOD schemas.
These cases are collected as unknown head and neck primary (C760), which will have no
evidence of primary tumor. This Schema ID was designed specifically to group together these

50 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
cases of an occult primary, a tumor that is not identified on physical exam or imaging
techniques.
o Note: Previous instructions were to code these types of cases to C148.
Coding Guidelines
This schema discriminator is based on several different criteria
Positive cervical lymph nodes. This is the overall term used for the head and neck regional lymph
nodes, which include Levels I-VII, and other group.
o For a complete listing of these lymph nodes, see AJCC 8
th
Edition Chapter 5: Staging
Head and Neck Cancer
o This same information can be found in the EOD and Summary Stage Manuals
No evidence of primary tumor and the suspected primary site is in the head and neck. A specific
head and neck primary site may be suspected, but if there is no clinical/pathological evidence of
the primary tumor, then the case is included in this schema
Epstein-Barr Virus (EBV): EBV positive cancers are associated with nasopharyngeal cancer.
o If the EBV (EBER) test is done and is positive, the primary site should be assigned to
C119 (nasopharynx, NOS) instead of C760, so that the Nasopharynx staging system can
be used. Nasopharynx has a T0, for no evidence of primary tumor
p16: p16 positive cancers in the head and neck are associated with oropharyngeal cancer. p16 is
a surrogate marker for Human Papilloma Virus (HPV).
o If the p16 test is done and positive (and EBV is negative or unknown), the primary site
should be assigned to C109 (oropharynx, NOS) instead of C760, so that the Oropharynx
staging system can be used. Oropharynx has a T0, for no evidence of primary tumor.
o Note: p16 is the only test that can be used for this discriminator. If there is another HPV
test that is positive, the p16 would still be negative for purposes of this data item.
Code 0: Not occult
Primary tumor is evident in the head and neck region; however, a specific primary site cannot be
identified.
Note 1: If overlapping lesions are evident and the primary site cannot be determined, this would
not be a C760, but C148 (overlapping lesions) (Schema ID 00118: Pharynx Other)
Note 2: For tumors in this category, C760 should be used sparingly. If C760 is assigned, this
would be collected in the following schemas: Schema ID 99999: Ill-Defined Other OR Schema ID
00450: Soft Tissue Other (if specified sarcoma)
o Examples include: Cases with limited information, historical case
Code 1: Occult, Negative cervical nodes (regional head and neck nodes)
No evidence of primary tumor or positive cervical lymph nodes (head and neck regional lymph
nodes), suspected head and neck primary.
This type of situation would be rare but would probably be diagnosed based on metastatic
disease, including distant lymph nodes (Mediastinal [excluding superior mediastinal node(s)])
This case would be collected in the Ill-Defined Other schema, or Soft Tissue Other (if specified
sarcoma).
Code 2: Not tested for EBV or p16 in head and neck regional nodes (EBV and p16 both unknown)
There is no documentation in the record regarding p16 or EBV.
Code 3: Unknown EBV, p16 negative in head and neck regional nodes

51 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
p16 is done and reported as negative. No documentation in the medical record regarding EBV.
Code 4: Unknown p16, EBV negative in head and neck regional nodes
EBV done and reported as negative. No documentation in the medical record regarding p16.
Code 5: Negative for both EBV and p16 in head and neck regional nodes
Both EBV and p16 done and reported as negative
EBV Positive EBV Negative EBV Unknown
p16 Positive C11.9 Nasopharynx
(00090:
Nasopharynx)
C10.9 Oropharynx
(00100: Oropharynx HPV-Mediated
(p16+))
C10.9 Oropharynx
(00100: Oropharynx HPV-Mediated
(p16+))
p16
Negative
C11.9 Nasopharynx
(00090:
Nasopharynx)
C76.0 Ill-Defined Site of the Head
and Neck
(00060: Cervical Lymph Nodes and
Unknown Primary)
C76.0 Ill-Defined Site of the Head
and Neck
(00060: Cervical Lymph Nodes and
Unknown Primary)
p16
Unknown
C11.9 Nasopharynx
(00090:
Nasopharynx)
C76.0 Ill-Defined Site of the Head
and Neck
(00060: Cervical Lymph Nodes and
Unknown Primary)
C76.0 Ill-Defined Site of the Head
and Neck
(00060: Cervical Lymph Nodes and
Unknown Primary)
Coding Instructions and Codes
Note 1: This schema discriminator is used to discriminate between head and neck tumors with unknown
primary site coded as C760. Some situations require that a more specific primary site be assigned.
00060: Cervical Lymph Nodes and Unknown Primary
Occult head and neck tumor with cervical metastasis in Levels I-VII, and other group lymph
nodes without a p16 immunostain or with negative results and without an Epstein-Barr virus
(EBV) encoded small RNAs (EBER) by in situ hybridization performed or with negative results are
staged using the AJCC Cervical Lymph Nodes and Unknown Primary Tumors of the Head and
Neck Staging System. Assign primary site C760; code the schema discriminator accordingly.
00090: Nasopharynx
Occult head and neck tumors with cervical metastasis in Levels I-VII, and other group lymph
nodes that is positive for Epstein–Barr virus (EBV+) (regardless of p16 status) encoded small
RNAs (EBER) identified by in situ hybridization are staged using the AJCC Nasopharynx Staging
System. Assign primary site C119; do NOT code this discriminator.
00100: Oropharynx HPV-Mediated (p16+)
Occult head and neck tumors with cervical metastasis in Levels I-VII, and other group lymph
nodes, p16 positive with histology consistent with HPV-mediated oropharyngeal carcinoma
(OPC), are staged using the AJCC HPV-Mediated (p16+) Oropharyngeal Cancer Staging System.
Assign primary site C109; do NOT code this discriminator

52 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
99999: Ill-Defined Other
If the tumor is not occult or does not have cervical metastasis in Levels I-VII, and other group
lymph nodes, it is not included in the AJCC Cervical Lymph Nodes and Unknown Primary Tumors
of the Head and Neck Staging System and will be classified as Ill-Defined Other for Summary
Staging
Note 2: If there is no evidence of the primary tumor, yet the physician “suspects” a specific head and
neck subsite, do not assign that primary site, but code C760 (see exceptions for EBV positive or p16
positive cancers.)
Code Description Schema ID #/Description
0 Not Occult 99999: Ill-Defined Other
00459: Soft Tissue Other (8941/3 only)
1 Occult, Negative cervical nodes (regional head
and neck nodes)
99999: Ill-Defined Other
00459: Soft Tissue Other (8941/3 only)
2 Not tested for EBV or p16 in head and neck
regional nodes (EBV and p16 both unknown)
00060: Cervical Lymph Nodes and
Unknown Primary
3 Unknown EBV, p16 negative in head and neck
regional nodes
00060: Cervical Lymph Nodes and
Unknown Primary
4 Unknown p16, EBV negative in head and neck
regional nodes
00060: Cervical Lymph Nodes and
Unknown Primary
5 Negative for both EBV and p16 in head and
neck regional nodes
00060: Cervical Lymph Nodes and
Unknown Primary
<Blank> Not C760, discriminator does not apply
Positive p16 in head and neck regional nodes,
EBV unknown or negative
Assign primary site C109
Positive EBV in head and neck regional nodes,
p16 positive, negative, or unknown
Assign primary site C119
Various
00010: Oropharynx HPV-Mediated (p16+)
00090: Nasopharynx
Return to Schema ID Table

53 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
Item Length: 1
NAACCR Item #: 3831
XML Parent-NAACCR ID: Tumor-extranodalExtensionHeadNeckClin
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00060: Cervical Lymph Nodes and Unknown Primary (2018+)
00071: Lip (2018+)
00072: Tongue Anterior (2018+)
00073: Gum (2018+)
00074: Floor of Mouth (2018+)
00075: Palate Hard (2018+)
00077: Mouth Other (2018+)
00080: Major Salivary Glands (2018+)
00090: Nasopharynx (2018+)
000100: Oropharynx HPV-Mediated (p16+)
00111: Oropharynx (p16-)
00112: Hypopharynx (2018+)
00121: Ethmoid Sinus (2018+)
00122: Nasal Cavity and Ethmoid Sinus (2018+)
00130: Larynx Other (2018+)
00131: Larynx Supraglottic (2018+)
00132: Larynx Glottic (2018+)
Description
Extranodal extension is defined as “the extension of a nodal metastasis through the lymph node capsule
into adjacent tissue” and is a prognostic factor for most head and neck tumors. This data item pertains
to clinical extension.
Rationale
Extranodal Extension Head and Neck Clinical is a Registry Data Collection Variable in AJCC. It was
previously collected as Head and Neck SSF #8 (Common SSF).
Definition
The presence of extranodal extension (ENE) from regional lymph nodes is an important prognostic factor
in some cancers because these patients are rarely cured without some type of systemic chemotherapy
or radiation. Extranodal extension is defined as metastatic tumor growing from within the lymph node
outward through the lymph node capsule and into surrounding connective tissues.
This data item for ENE detected clinically.
Coding guidelines
Code 0 when there are positive nodes clinically, but ENE not identified/not present.

54 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code 1 when there are positive nodes clinically, ENE is identified by physical exam WITH or
WITHOUT imaging
Code 2 when there are positive nodes clinically, ENE is identified by biopsy (microscopically
confirmed)
Code 4 when there are positive nodes clinically, ENE is identified, but not known how identified
Code 7 when nodes are clinically negative (cN0)
Code 9 when
o No information in the medical record
o Positive nodes clinically, not evaluated (assessed) for ENE
o Positive nodes clinically, unknown if evaluated (assessed) for ENE
o Lymph nodes not evaluated (assessed) clinically
o Unknown if lymph nodes evaluated (assessed) clinically
Additional Information
Source documents: pathology report, imaging reports, physical exam
Other names: ENE, extracapsular extension, ECE
Coding Instructions and Codes
Note 1: Physician statement of extranodal extension (ENE) clinically or physician clinical stage indicating
the absence or presence of ENE can be used to code this data item when no other information is
available. Physical exam alone is sufficient to determine Clinical ENE.
Note 2: The assessment of ENE must be based on evidence acquired prior to definitive surgery of the
primary site, chemotherapy, radiation or other type of treatment, i.e., the clinical timeframe for staging.
The assessment for ENE in addition to physical examination may include imaging, biopsy of the
regional lymph node, and/or biopsy of tissues surrounding the regional lymph node.
Imaging alone is not enough to determine or exclude ENE.
Note 3: Be aware that the rules for coding ENE for head and neck sites compared to non-head and neck
sites are different.
Note 4: Code 0 when lymph nodes are determined to be clinically positive and physical examination
does not indicate any signs of extranodal extension.
Note 5: Code 1 when
ENE is unquestionable as determined by physical examination
o Clinical ENE is described in the AJCC Head and Neck Staging System as “Unambiguous
evidence of gross ENE on clinical examination (e.g., invasion of skin, infiltration of
musculature, tethering to adjacent structures, or cranial nerve, brachial plexus,
sympathetic trunk, or phrenic nerve invasion with dysfunction)”
The terms ‘fixed’ or ‘matted’ are used to describe lymph nodes
Other terms for ENE include: ‘extranodal spread’, ‘extracapsular extension’, or ‘extracapsular
spread’.
Note 6: Code 9 when physical exam is not available AND at least one of the following
No additional information
Statement of lymph node involvement with no information on ENE

55 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Lymph node biopsy (e.g., FNA, core, incisional, excisional, sentinel node) performed and is
negative for ENE or not stated
Code Description
0 Regional lymph nodes involved, ENE not present/not identified during diagnostic workup
1 Regional lymph nodes involved, ENE present/identified during diagnostic workup, based on
physical exam WITH or WITHOUT imaging
2 Regional lymph nodes involved, ENE present/identified during diagnostic workup, based on
microscopic confirmation
4 Regional lymph nodes involved, ENE present/identified, unknown how identified
7 No lymph node involvement during diagnostic workup (cN0)
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error)
9 Not documented in medical record
ENE not assessed during diagnostic workup, or unknown if assessed
Clinical assessment of lymph nodes not done, or unknown if done
Return to Schema ID Table

56 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3832: Extranodal Extension Head and Neck Pathological
Item Length: 3
NAACCR Item #: 3832
XML Parent-NAACCR ID: Tumor-extranodalExtensionHeadNeckPath
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00060: Cervical Lymph Nodes and Unknown Primary (2018+)
00071: Lip (2018+)
00072: Tongue Anterior (2018+)
00073: Gum (2018+)
00074: Floor of Mouth (2018+)
00075: Palate Hard (2018+)
00077: Mouth Other (2018+)
00080: Major Salivary Glands (2018+)
00090: Nasopharynx (2018+)
000100: Oropharynx HPV-Mediated (p16+)
00111: Oropharynx (p16-)
00112: Hypopharynx (2018+)
00121: Ethmoid Sinus (2018+)
00122: Nasal Cavity and Ethmoid Sinus (2018+)
00130: Larynx Other (2018+)
00131: Larynx Supraglottic (2018+)
00132: Larynx Glottic (2018+)
Description
Extranodal extension (ENE) is defined as “the extension of a nodal metastasis through the lymph node
capsule into adjacent tissue” and is a prognostic factor for most head and neck tumors. This data item
pertains to pathological staging extension.
Rationale
Extranodal Extension Head and Neck Pathological is a Registry Data Collection Variable in AJCC. It was
previously collected as Head and Neck SSF #9 (Common SSF).
Definition
The presence of extranodal extension (ENE) from regional lymph nodes is an important prognostic factor
in some cancers because these patients are rarely cured without some type of systemic chemotherapy
or radiation. Extranodal extension is defined as metastatic tumor growing from within the lymph node
outward through the lymph node capsule and into surrounding connective tissues.
This data item for ENE that is detected pathologically for head and neck primaries.
Coding guidelines
Code 0.0 when there are positive nodes pathologically, but ENE not identified/not present.

57 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code the actual size of the ENE in the range 0.1-9.9 mm
Code X.1 when actual size of the ENE is 10 mm or greater
Code X.2 when stated to be microscopic [ENE (mi)]
Code X.3 when stated to be major [ENE (ma)]
Code X.4 when size not documented, unknown whether microscopic (mi) or major (ma)
Code X.7 when nodes are surgically resected, and they are negative (pN0)
Code X.9 when
o No information in the medical record
o Positive nodes pathologically, not evaluated (assessed) for ENE
o Positive nodes pathologically, unknown if evaluated (assessed) for ENE
o Lymph nodes not evaluated (assessed) pathologically (no surgical resection of lymph
nodes)
o Unknown if lymph nodes evaluated pathologically (assessed)
Additional Information
Source documents: pathology report from surgical resection
Other names: ENE, extracapsular extension, ECE
Coding Instructions and Codes
Note 1: Physician statement of extranodal extension (ENE) pathologically during a lymph node
dissection or physician pathological stage indicating the absence or presence of ENE can be used to code
this data item when no other information is available.
Note 2: Extranodal extension is defined as “the extension of a nodal metastasis through the lymph node
capsule into adjacent tissue.” ENE is the preferred terminology. Other names include: extranodal spread,
extracapsular extension, or extracapsular spread.
“A regional node extending into a distant structure or organ is categorized as ENE and is not
recorded as distant metastatic disease.”
Note 3: Code the status of ENE assessed on histopathologic examination of surgically resected involved
regional lymph node(s). Do not code ENE from a lymph node biopsy (FNA, core, incisional, or the
absence of ENE from a sentinel). Do not code ENE for any distant lymph nodes. Code the status of ENE
based on the following criteria
Code 0.0
o Absence of ENE, positive lymph nodes assessed by lymph node dissection
o 1292: Scope of Regional Lymph Node Surgery must be 3-7
Codes 0.1-9.9, X.1, X.2, X.3, X.4 as appropriate for
o Presence of ENE assessed by Sentinel Lymph Node biopsy
o Presence of ENE assessed by lymph node biopsy
o 1292: Scope of Regional Lymph Node Surgery must be 2-7
Code X.7 as appropriate for
o Lymph nodes negative for cancer assessed by Sentinel lymph node biopsy or lymph
node dissection
o 1292: Scope of Regional Lymph Node Surgery must be 2-7
Code X.9
o Absence of ENE, positive lymph nodes assessed by Sentinel Lymph Node biopsy

58 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
A positive Sentinel Lymph Node biopsy cannot assess the absence of ENE, only
the presence of it. This is because there is not enough surrounding tissue in a
Sentinel Lymph node biopsy to accurately assess ENE
If codes 0.1-0.9, X.1-X.7 are used, this indicates that the lymph nodes were surgically resected or
a Sentinel Lymph Node biopsy was done and Scope of Regional Lymph Node Surgery [NAACCR
Data Item: 1292] must be 2-7
Note 4: Be aware that the rules for coding ENE for head and neck sites compared to non-head and neck
sites are different.
Note 5: Definitions of ENE subtypes and rules:
Microscopic ENE [ENE (mi)] is defined as less than or equal to 2 mm.
Major ENE [ENE (ma)] is defined as greater than 2 mm.
Both ENE (mi) and ENE (ma) qualify as ENE (+) for definition of pN.
Note 6: The measurement of ENE is the distance from the lymph node capsule in millimeters (mm).
Code Description
0.0 Lymph nodes positive for cancer but ENE not identified or negative
0.1-9.9 ENE 0.1 to 9.9 mm
X.1 ENE 10 mm or greater
X.2 ENE microscopic, size unknown
Stated as ENE (mi)
X.3 ENE major, size unknown
Stated as ENE (ma)
X.4 ENE present, microscopic or major unknown, size unknown
X.7 Surgically resected regional lymph nodes negative for cancer (pN0)
X.8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X.8 may result in an edit error)
X.9 Not documented in medical record
No surgical resection of regional lymph nodes
ENE not assessed pathologically, or unknown if assessed
Pathological assessment of lymph nodes not done, or unknown if done
Return to Schema ID Table

59 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other)
For head and neck sites, regional lymph node information is coded in several different data items.
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
The Head and Neck Levels are defined as
1. Level I is subdivided into levels IA and IB, which contain the submental and submandibular
triangles bounded by the anterior and posterior bellies of the digastric muscle, the hyoid bone
inferiorly, and the body of the mandible superiorly. Lymph node chains at this level:
Submental (Level IA), submandibular (Level IB), submaxillary (Level IB)
2. Level II is subdivided into levels IIA and IIB, which contain the upper jugular lymph nodes and
extend from the level of the skull base superiorly to the hyoid bone inferiorly. A vertical plane
defined by the spinal accessory nerve is the boundary between level IIA (anterior to spinal
accessory nerve) and IIB (posterior to spinal accessory nerve). Lymph node chains at this level:
Jugulodigastric (subdigastric), upper deep cervical, upper jugular
3. Level III contains the middle jugular lymph nodes from the hyoid bone superiorly to the level of
the lower border of the cricoid cartilage inferiorly. Lymph node chains at this level:
Middle deep cervical, mid-jugular
4. Level IV contains the lower jugular lymph nodes from the level of the cricoid cartilage superiorly
to the clavicle inferiorly. Lymph node chains at this level:
Jugulo-omohyoid (supraomohyoid), lower deep cervical, lower jugular
5. Level V is subdivided into levels VA and VB, which contain the lymph nodes in the posterior
triangle bounded by the anterior border of the trapezius muscle posteriorly, the posterior
border of the sternocleidomastoid muscle anteriorly, and the clavicle inferiorly. For descriptive
purposes, Level V may be further subdivided into upper (VA) and lower (VB) levels
corresponding to a plane defined by the inferior border of the cricoid cartilage. Lymph node
chains at this level:
Posterior cervical, posterior triangle (spinal accessory, transverse cervical [upper,
middle, and lower, corresponding to the levels that define upper, middle, and lower
jugular nodes]), supraclavicular
6. Level VI contains the lymph nodes of the anterior central compartment from the hyoid bone
superiorly to the suprasternal notch inferiorly. On each side, the lateral boundary is formed by
the medial border of the carotid sheath. Lymph node chains at this level:
Laterotracheal, Paralaryngeal, paratracheal (above suprasternal notch), perithyroidal,
Precricoid (Delphian), Prelaryngeal, recurrent laryngeal)
7. Level VII contains the lymph nodes inferior to the suprasternal notch in the superior
mediastinum. Lymph node chains at this level:
Esophageal groove, paratracheal (below suprasternal notch), Pretracheal (below
suprasternal notch)

60 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
8. Other head and neck lymph nodes:
Cervical, NOS; deep cervical (NOS), facial, buccinator (buccal), infraauricular, internal
jugular (NOS), intraparotid, mandibular, nasolabial, parapharyngeal, parotid,
periparotid, preauricular, retroauricular (mastoid), retropharyngeal, suboccipital
Coding guidelines
Example 1: A carcinoma of the base of tongue involves bilateral submandibular nodes and left
upper, mid-, and lower jugular nodes, the largest measuring 4 cm. There is no extracapsular
extension. These are level I, II, III, and IV lymph nodes according to AJCC definitions.
Levels I-III: Code 7 (Levels I, II and III lymph nodes involved) to show that levels I, II, and
III are involved
Levels IV-V: Code 1 to show that level IV is involved
Levels VI-VII: Code 0 for no other nodes involved
Head and Neck, Other: Code 0 for no other nodes involved
Example 2: Patient diagnosed elsewhere with carcinoma of oropharynx with cervical lymph
node involvement. No other information available. All Head and Neck Level data items are
coded to 9 since there is no specific information about the levels.
Levels I-III: Code 9
Levels IV-V: Code 9
Levels VI-VII: Code 9
Head and Neck, Other: Code 9
Coding NOS
Note: When the only information available is “Regional nodes, NOS” or “Cervical nodes, NOS” or
“Internal jugular nodes, NOS” or “Lymph nodes, NOS,” code 9. In other words, if regional nodes
are known to be positive but the level(s) of nodes involved is unknown, code 9.
Coding a Node That Overlaps Two Levels
Note: If a lymph node is described as involving two levels, code both levels.
o Example: Physical examination for a floor of mouth cancer describes a large lymph node
mass low in level II stretching into Level III. Code 6 for Levels II-III because both Level II
and Level III are mentioned.

61 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3876: LN Head and Neck Levels I-III
Item Length: 1
NAACCR Item #: 3876
XML Parent-NAACCR ID: Tumor-lnHeadAndNeckLevels1To3
NAACCR Alternate Name: Lymph Nodes Head and Neck Levels I-III
Active years: 2018+
Schemas(s):
00060: Cervical Lymph Nodes and Unknown Primary (2018+)
00140: Melanoma Head and Neck (2018+)
Description
Lymph Nodes for Head and Neck, Levels I-III records the involvement of Levels I-III lymph nodes.
Rationale
Level of nodal involvement is a Registry Data Collection Variable in AJCC for several head and neck
chapters. This data item was previously collected as Head and Neck SSF #3 (common SSF).
Definition
This data item is used to code the presence or absence of lymph node involvement in head and neck
levels I-III. The definitions of the levels are the same for all applicable head and neck sites.
Note: This data item was previously collected for all head and neck sites. It is now only clinically
relevant for unknown head and neck primaries with positive cervical (head and neck) lymph
nodes and mucosal melanomas of the head and neck.
See 3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Levels I-III lymph node involvement can be used to code this data item
when no other information is available.
Note 2: Head and Neck Lymph Node Involvement is coded in the following data items
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
Note 3: Code the presence or absence of lymph node involvement for Levels I-III.
For more information on Levels I-III lymph nodes, see AJCC 8
th
edition, Chapter 5: Staging Head
and Neck Cancers, Table 5.1
Note 4: Pathological information takes priority over clinical.
Note 5: If involved regional node levels are documented as a range, or if the involved nodes overlap
multiple levels, code all levels specified.

62 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 6: If information is available on some nodes, but the others are unknown, code what is known.
Example: Multiple lymph nodes involved, level II documented, but the other levels not
mentioned. Code 2 to indicate level II involvement.
Code Description
0 No involvement in Levels I, II, or III lymph nodes
1
Level I lymph node(s) involved
2 Level II lymph node(s) involved
3 Level III lymph node(s) involved
4 Levels I and II lymph nodes involved
5 Levels I and III lymph nodes involved
6 Levels II and III lymph nodes involved
7 Levels I, II and III lymph nodes involved
8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error)
9 Not documented in medical record
Positive nodes, but level of positive node(s) unknown
Lymph node levels I-III not assessed, or unknown if assessed
Return to Schema ID Table

63 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3877: LN Head and Neck Levels IV-V
Item Length: 1
NAACCR Item #: 3877
XML Parent-NAACCR ID: Tumor-lnHeadAndNeckLevels4To5
NAACCR Alternate Name: Lymph Nodes Head and Neck Levels IV-V
Active years: 2018+
Schemas(s):
00060: Cervical Lymph Nodes and Unknown Primary (2018+)
00140: Melanoma Head and Neck (2018+)
Description
Lymph Nodes for Head and Neck, Levels IV-V records the involvement of Levels IV-V lymph nodes.
Rationale
Level of nodal involvement is a Registry Data Collection Variable in AJCC. This data item was previously
collected as Head and Neck SSF #4 (common SSF).
Definition
This data item is used to code the presence or absence of lymph node involvement in head and neck
levels IV-V. The definitions of the levels are the same for all applicable head and neck sites.
Note: This data item was previously collected for all head and neck sites. It is now only clinically
relevant for unknown head and neck primaries with positive cervical (head and neck) lymph
nodes and mucosal melanomas of the head and neck.
See 3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Levels IV-V lymph node involvement can be used to code this data item
when no other information is available.
Note 2: Head and Neck Lymph Node Involvement is coded in the following data items
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
Note 3: Code the presence or absence of lymph node involvement for Levels IV-V.
For more information on Levels IV-V lymph nodes, see AJCC 8
th
edition, Chapter 5: Staging Head
and Neck Cancers, Table 5.1
Note 4: If lymph nodes are described only as “supraclavicular,” try to determine if they are in Level IV
(deep to the sternocleidomastoid muscle, in the lower jugular chain) or Level V (in the posterior triangle,
inferior to the transverse cervical artery) and code appropriately.

64 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
If the specific level cannot be determined, or is documented as supraclavicular with no further
information, code them as Level V nodes
Note 5: Pathological information takes priority over clinical.
Note 6: If involved regional node levels are documented as a range, and/or if the involved nodes overlap
multiple levels, code all levels specified.
Note 7: If information is available on some nodes, but the others are unknown, code what is known.
Example: Multiple lymph nodes involved, level V documented, but the other levels not
mentioned. Code 2 to indicate level V involvement.
Code Description
0 No involvement in Levels IV or V lymph nodes
1 Level IV lymph node(s) involved
2 Level V lymph node(s) involved
3 Levels IV and V lymph node(s) involved
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error)
9 Not documented in medical record
Positive nodes, but level of positive node(s) unknown
Lymph node levels IV-V not assessed, or unknown if assessed
Return to Schema ID Table

65 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3878: LN Head and Neck Levels VI-VII
Item Length: 1
NAACCR Item #: 3878
XML Parent-NAACCR ID: Tumor-lnHeadAndNeckLevels6To7
NAACCR Alternate Name: Lymph Nodes Head and Neck Levels VI-VII
Active years: 2018+
Schemas(s):
00060: Cervical Lymph Nodes and Unknown Primary
00140: Melanoma Head and Neck
Description
Lymph Nodes for Head and Neck, Levels VI-VII records the involvement of Levels VI-VII lymph nodes.
Rationale
Level of nodal involvement is a Registry Data Collection Variable in AJCC. This data item was previously
collected as Head and Neck SSF #5 (common SSF).
Definition
This data item is used to code the presence or absence of lymph node involvement in head and neck
levels VI and VII. The definitions of the levels are the same for all applicable head and neck sites.
Note 1: This data item was previously collected for all head and neck sites. It is now only
clinically relevant for unknown head and neck primaries with positive cervical (head and neck)
lymph nodes and mucosal melanomas of the head and neck.
Note 2: In Collaborative Stage v2 (CSv2), Facial Lymph Nodes were collected with Levels VI-VII.
They have now been moved to the “other” group.
See 3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Levels VI-VII lymph node involvement can be used to code this data item
when no other information is available.
Note 2: Head and Neck Lymph Node Involvement is coded in the following data items
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
Note 3: Code the presence or absence of lymph node involvement for Levels VI-VII.
For more information on Levels VI-VII lymph nodes, see AJCC 8
th
edition, Chapter 5: Staging
Head and Neck Cancers, Table 5.1
Note 4: Pathological information takes priority over clinical.

66 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 5: If involved regional node levels are documented as a range, or if the involved nodes overlap
multiple levels, code all levels specified.
Note 6: If information is available on some nodes, but the others are unknown, code what is known.
Example: Multiple lymph nodes involved, level VI documented, but the other levels not
mentioned. Code 1 to indicate level VI involvement.
Code Description
0 No involvement in Levels VI or VII lymph nodes
1 Level VI lymph node(s) involved
2 Level VII lymph node(s) involved
3 Levels VI and VII lymph node(s) involved
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error)
9 Not documented in medical record
Positive nodes, but level of positive node(s) unknown
Lymph nodes levels VI-VII not assessed, or unknown if assessed
Return to Schema ID Table

67 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3879: LN Head and Neck Other
Item Length: 1
NAACCR Item #: 3879
XML Parent-NAACCR ID: Tumor-lnHeadAndNeckOther
NAACCR Alternate Name: Lymph Nodes Head and Neck Other
Active years: 2018+
Schemas(s):
00060: Cervical Lymph Nodes and Unknown Primary
00140: Melanoma Head and Neck
Description
Lymph Nodes for Head and Neck, Other records the involvement of lymph nodes other than Levels I-III,
IV-V, and VI-VII.
Rationale
Level of nodal involvement is a Registry Data Collection Variable in AJCC. This data item was previously
collected as Head and Neck SSF #6 (common SSF).
Definition
This data item is used to code the presence or absence of lymph node involvement for other head and
neck lymph nodes. The definitions of the levels are the same for all applicable head and neck sites.
Note 1: This data item was previously collected for all head and neck sites. It is now only
clinically relevant for unknown head and neck primaries with positive cervical (head and neck)
lymph nodes and mucosal melanomas of the head and neck.
Note 2: In Collaborative Stage v2 (CSv2), Facial Lymph Nodes were collected with Levels VI-VII.
They are now collected with the “other” lymph nodes.
See 3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of other head and neck lymph node involvement can be used to code this
data item when no other information is available.
Note 2: Head and Neck Lymph Node Involvement is coded in the following data items
3876: LN Head and Neck Levels I-III
3877: LN Head and Neck Levels IV-V
3878: LN Head and Neck Levels VI-VII
3879: LN Head and Neck Other
Note 3: Code the presence or absence of lymph node involvement for the “other” group.
For more information on the other head and neck lymph nodes, see AJCC 8
th
edition, Chapter 5:
Staging Head and Neck Cancers, Table 5.1

68 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: Pathological information takes priority over clinical.
Note 5: If involved regional node levels are documented as a range, and/or if the involved nodes overlap
multiple levels, code 7.
Note 6: If information is available on some nodes, but the others are unknown, code what is known.
Example: Multiple lymph nodes involved, preauricular documented, but the other levels not
mentioned. Code 4 to indicate preauricular involvement.
Code Description
0 No involvement in other head and neck lymph node regions
1 Buccinator (facial) lymph node(s) involved
2 Parapharyngeal lymph node(s) involved
3 Periparotid and intraparotid lymph node(s) involved
4 Preauricular lymph node(s) involved
5
Retropharyngeal lymph node(s) involved
6
Suboccipital/retroauricular lymph node(s) involved
7 Any combination of codes 1-6
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Positive nodes, but level of positive node(s) unknown
Other Head and Neck lymph nodes not assessed, or unknown if assessed
Return to Schema ID Table

69 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3883: LN Size
Item Length: 4
NAACCR Item #: 3883
XML Parent-NAACCR ID: Tumor-lnSize
NAACCR Alternate Name: Lymph Nodes Size
Active years: 2018+
Schema(s):
00060: Cervical Lymph Nodes and Unknown Primary
00071: Lip
00072: Tongue Anterior
00073: Gum
00074: Floor of Mouth
00075: Palate Hard
00076: Buccal Mucosa
00077: Mouth Other
00080: Major Salivary Glands
00090: Nasopharynx
00100: Oropharynx HPV-Mediated (p16+)
00111: Oropharynx (p16-)
00112: Hypopharynx
00121: Maxillary Sinus
00122: Nasal Cavity and Ethmoid Sinuses
00130: Larynx Other
00131: Larynx SupraGlottic
00132: Larynx Glottic
00133: Larynx SubGlottic
00140: Melanoma Head and Neck
00150: Cutaneous Carcinoma of Head and Neck
Description
Lymph Nodes Size records diameter of the involved regional lymph node(s) with the largest diameter of
any involved regional lymph node(s). Pathological measurement takes precedence over a clinical
measurement for the same node.
Rationale
Lymph Nodes Size is a Registry Data Collection Variable in AJCC for several chapters. It was previously
collected in the Head and Neck chapters as Size of Lymph Nodes, SSF #1.
Definition
This data item is used to code the size of involved lymph nodes and is recorded in millimeters.
Coding guidelines
Code the largest diameter of any involved regional lymph nodes for head and neck (cervical lymph
nodes). The measurement can be pathological, if available, or clinical.
Code 0.0 when no regional lymph nodes are involved
Code XX.1 for 100 millimeters (10 cm) or greater

70 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code XX.2 for microscopic focus or foci only and no size of focus given
Code XX.3 for lymph node met less than 1 cm (10 mm)
o Lymph node described as “subcentimeter)
Code XX.9 when
o Positive lymph nodes but size not stated
o No information about regional lymph nodes
o Lymph nodes not assessed or unknown if assessed
In order to align with the CAP guidelines, additional codes have been added for “at least” categories
which are used in the CAP protocols. Only use these codes when the pathologist has used this
terminology to indicate the lymph node size.
XX.4: Describes a lymph node size at least 2 cm (20 mm)
XX.5: Described a lymph node size at least 3 cm (30 mm)
XX.6: Describes a lymph node size at least 4 cm (40 mm)
XX.7: Describes a lymph node size 5 cm (50 mm) or greater
Coding Instructions and Codes
Note 1: Physician statement of Lymph Nodes Size can be used to code this data item when no other
information is available.
Note 2: If the same largest involved node (or same level) is examined both clinically and pathologically,
record the size of the node from the pathology report, even if it is smaller.
Example: Clinical evaluation shows 1.5 cm (15 mm) Level II lymph node, pathological
examination shows Level II 1.3 cm (13 mm). Code 13.0.
Note 3: If the largest involved node is not examined pathologically, use the clinical node size.
Note 4: Do not code the size of any distant nodes.
Code Description
0.0 No regional lymph node involvement
Non-invasive neoplasm (behavior /2)
0.1-99.9 0.1 – 99.9 millimeters (mm)
(Exact size of lymph node to nearest tenth of a mm)
XX.1 100 millimeters (mm) or greater
XX.2 Microscopic focus or foci only and no size of focus given
XX.3 Described as "less than 1 centimeter (cm)" or “subcentimeter”
XX.4 Described as "at least” 2 cm
XX.5 Described as "at least” 3 cm
XX.6 Described as "at least” 4 cm
XX.7 Described as greater than 5 cm
XX.8
Not applicable: Information not
collected for this case
(If this item is required by your standard setter, use of code XX.8 will result in an edit error)
XX.9 Not documented in medical record
Regional lymph node(s) involved, size not stated
Lymph Nodes Size not assessed, or unknown if assessed
Return to Schema ID Table

71 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00071: Lip (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00072: Tongue Anterior (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00073: Gum (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00074: Floor of Mouth (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00075: Palate Hard (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size

72 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00076: Buccal Mucosa (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00077: Mouth Other (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00080: Major Salivary Glands (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00090: Nasopharynx (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
Return to Schema ID Table

73 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00090: Nasopharynx (2018+)
3926: Schema Discriminator 1: Nasopharynx/Pharyngeal Tonsil
Primary site C111 only
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00090: Nasopharynx
00100: Oropharynx HPV-Mediated (p16+)
00111: Oropharynx (p16-)
Definition
Nasopharynx and pharyngeal tonsil have the same ICD-O topography code (C111). However, for
purposes of stage grouping AJCC 8
th
edition, nasopharynx and pharyngeal tonsil are staged in different
chapters. A schema discriminator is necessary to distinguish between these primary sites so that the
appropriate chapter/schema is used.
Coding Instructions and Codes
Note: A schema discriminator is used to discriminate for primary site C111: Posterior wall of
nasopharynx. Code the specific site in which the tumor arose.
00090: Nasopharynx (see code 1)
Used to stage for the following primary site description: posterior wall of nasopharynx (NOS)
00100 (Oropharynx HPV-Mediated (p16+)) or 00111 (Orpharynx (p16-)) (see code 2)
Oropharynx Staging Systems are used for the following primary site descriptions. An additional
schema discriminator will be used to distinguish between the AJCC HPV-Mediated (p16+)
Oropharyngeal Cancer and Oropharynx (p16-) and Hypopharynx Staging System
o Adenoid
o Pharyngeal tonsil
Code Description Schema ID #/Description
1 Posterior wall of nasopharynx, NOS 00090: Nasopharynx
2 Adenoid
Pharyngeal tonsil
3927: Schema discriminator 2:
Oropharyngeal p16
<Blank> Primary Site is NOT C111, Discriminator is not necessary
Return to Schema ID Table

74 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00100: Oropharynx HPV-Mediated (p16+) (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
See 00090: Nasopharynx (2018+)
3926: Schema Discriminator 1: Nasopharynx/Pharyngeal Tonsil

75 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00100: Oropharynx HPV-Mediated (p16+) (2018+)
3927: Schema Discriminator 2: Oropharyngeal p16
Item Length: 1
NAACCR Item #: 3927
XML Parent-NAACCR ID: Tumor-schemaDiscriminator2
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00100: Oropharynx HPV-Mediated (p16+)
00111: Oropharynx (p16-)
Definition
Staging for oropharyngeal cancers changed in the AJCC 8
th
edition. Chapter 10 is now for p16+ tumors,
while Chapter 11 is for p16- negative tumors or where the p16 is not assessed or unknown. A schema
discriminator is necessary to determine the p16 status so that the appropriate chapter/schema is used.
Coding Instructions and Codes
Note 1: A schema discriminator is used to discriminate between oropharyngeal tumors that are p16
positive and oropharyngeal tumors that are p16 negative OR p16 status unknown.
Note 2: Only the HPV p16+ test can be used. If another HPV test is done, code 9.
00100: Oropharynx HPV-Mediated (p16+) (see code 2)
o Used for p16 (+) (positive)
00111: Oropharynx (p16-)
o p16 expression of weak intensity or limited distribution (see code 1)
o p16 without an immunostain performed (see code 9)
Code
Description
Schema ID#/Description
1 p16 Negative; Nonreactive 00111: Orpharynx (p16-)
2 p16 Positive; HPV Positive; Diffuse, Strong reactivity 00100: Oropharynx HPV-Mediated
(p16+)
9 Not tested for p16; Unknown 00111: Orpharynx (p16-)
Return to Schema ID Table

76 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00111: Oropharynx (p16-) (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
See 00090: Nasopharynx (2018+)
3926: Schema Discriminator 1: Nasopharynx/Pharyngeal Tonsil
See 00100: Oropharynx HPV-Mediated (p16+)
3927: Schema Discriminator 2: Oropharyngeal p16
Return to Schema ID Table

77 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00112: Hypopharynx (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00121: Maxillary Sinus (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00122: Nasal Cavity and Ethmoid Sinuses (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00130: Larynx Other (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00131: Larynx Supraglottic (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00132: Larynx Glottic (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size

78 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00133: Larynx SubGlottic (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3883: LN Size
00140: Mucosal Melanoma of the Head and Neck (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3831: Extranodal Extension Head and Neck Clinical
3832: Extranodal Extension Head and Neck Pathological
3876-3879: Head and Neck Regional Lymph Nodes (Levels I-VII, Other)
o 3876: LN Head and Neck Levels I-III
o 3877: LN Head and Neck Levels IV-V
o 3878: LN Head and Neck Levels VI-VII
o 3879: LN Head and Neck Other
00150: Cutaneous Carcinoma of the Head and Neck (2018+)
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3883: LN Size
See 00200: Colon and Rectum for the following data item
3909: Perineural Invasion
Return to Schema ID Table

79 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00150: Cutaneous Carcinoma of the Head and Neck (2018+)
3858: High Risk Histologic Features
Item Length: 1
NAACCR Item #: 3858
XML Parent-NAACCR ID: Tumor-highRiskHistologicFeatures
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00150: Cutaneous Carcinoma of Head and Neck (2018+)
Description
High Risk Histologic Features are defined in AJCC 8 Chapter 15 to include the terms “poor
differentiation, desmoplasia, sarcomatoid differentiation, undifferentiated.” High risk histologic
features are a prognostic factor for cutaneous cell carcinomas of the head and neck.
Rationale
High Risk Histologic Features is a Registry Data Collection Variable in AJCC. It was previously collected as
Skin, CS SSF # 12.
Definition
In addition to the tumor size (diameter, not depth), the presence of certain specific high-risk features is
of prognostic significance for skin cancers of the head and neck.
In Collaborative Stage v2 (CSv2), which was based on AJCC 7
th
edition, the number of high risk features
impacted the assignment of T. This is no longer the case. The type of high risk feature is now recorded
instead of the number.
Coding guidelines
Record the presence of high-risk features
Code 1 for desmoplasia
Code 2 for poor differentiation (grade 3)
Code 3 for sarcomatoid differentiation (features)
Code 4 for undifferentiated (grade 4)
Code 5 when more than one feature is present
Code 6 when high risk features are present, but it is not specified which one
Code 9 when
o Not documented in medical record
o High risk features not evaluated (assessed)
o Unknown if high-risk features evaluated (assessed)
Additional Information
Source documents: pathology report, consultation report, other statements in the medical
record

80 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Other names: high risk histologic features, high risk tumor features
Coding Instructions and Codes
Note 1: Physician statement of high risk histologic features can be used to code this data item when no
other information is available.
Note 2: High risk histologic features include
Desmoplasia
Poor differentiation (grade 3)
Sarcomatoid differentiation (features)
Undifferentiated (grade 4)
Note 3: Code the presence or absence of high risk histologic features as documented in the pathology
report.
Note 4: Code 5 if more than one high risk histologic feature is present.
Code Description
0
No high risk histologic features
Non-invasive neoplasm (behavior /2)
1 Desmoplasia
2 Poor differentiation (grade 3)
3 Sarcomatoid differentiation
4 Undifferentiated (grade 4)
5 Multiple high risk histologic features
6 Histologic features, NOS (type of high risk histologic feature not specified)
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error)
9
Not documented in medical record
High risk histologic features not assessed or unknown if assessed
Return to Schema ID Table
81 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
GASTROINTESTINAL TRACT (UPPER AND LOWER)

82 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00161: Esophagus (including GE junction) Squamous (2018+)
3926: Schema Discriminator 1: EsophagusGEJunction (EGJ)/Stomach
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00161: Esophagus (including GE junction) Squamous (2018+)
00169: Esophagus (including GE junction) (excluding Squamous) (2018+)
00170: Stomach (2018+)
Definition
The esophagus chapter of the AJCC Cancer Staging Manual 8
th
edition includes the esophagogastric
junction (also called the cardia or gastroesophageal junction) and the proximal 2 cm of the stomach. The
cardia is defined as the opening or junction between the esophagus and the stomach, and it is between
0.1 and 0.4 cm in length. This 2-cm boundary measurement is based on the Siewert classification of
gastroesophageal cancers, which defines an area 2 cm above and 2 cm below the cardia or
esophagogastric junction. Both of these areas are coded to primary site C160, so a discriminator is
needed to get to the correct chapter.
Note: This is different from AJCC 7
th
edition (CSv2) where the measurement was 5 cm.
To determine whether a cancer of the cardia should be coded according to the esophagus schema or the
stomach system, it is necessary to identify the midpoint or epicenter of the tumor. If the midpoint is at
or above the cardia, the tumor is esophageal. If the midpoint of the tumor is within 2 cm distal to the
gastroesophageal junction (GEJ) and the lesion extends to or across the GEJ, the case should be coded
with the esophagus system. If the midpoint of the tumor is within 2 cm distal to the GEJ and the lesion
does not extend to the GEJ, the case should be coded with the stomach schema. Any tumor with a
midpoint more distal than 2 cm from the GEJ is coded with the stomach schema.
Select the code that best describes the location and extent of the tumor, and the computer
algorithm will bring the correct schema to the screen
Coding Notes and Instructions
Note 1: Under primary site code C160, there are two different structures that are staged differently.
Esophagogastric junction (Esophagus Schemas)
Cardia of the Stomach (Stomach schema)
Note 2: The gastroesophageal junction (GEJ) (primary site C160) is a poorly defined anatomic area that
represents the junction between the distal esophagus and the proximal stomach (cardia). The true
anatomic GEJ corresponds to the most proximal aspect of the gastric folds, which represents an
endoscopically apparent transition point in most individuals.

83 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 3: The cardia (also assigned primary site C160) is the first part of the stomach. It is the region
where the stomach meets the end of the esophageal tube. This region is also referred to as the Z-line or
the esophagogastric junction.
Note 4: Physician's statement can be used to code this data item when no other information is available.
Example: Patient diagnosed with tumor involving the cardia. No other information available.
Physician stages the patient using the Esophagus Staging System/CAP protocol
o Answer: Code 2 based on physician using the Esophagus Staging System
Note 5: Tumors with their midpoint (epicenter) in the GE Junction are staged as Esophagus, while
tumors with their midpoint (epicenter) in the cardia/stomach are staged using the Stomach Staging
System.
Note: The CAP protocol uses "midpoint" instead of "epicenter." This is the pathologist's
assessment of the point of tumor origin, regardless of tumor extension into other tissues.
Chapter 16: Esophagus and Esophagogastric Junction (see code 2)
Note 1: Use code 2 when
EGJ is documented as involved and the midpoint (epicenter) is within the proximal (above) 2 cm
of the cardia
EGJ is documented as involved and there is no mention of extension into the stomach or
stomach involvement
Example 1: MRI: Findings most consistent with metastatic GE junction cancer. Upper EUS:
Medium-sized, fungating, polypoid and ulcerated mass with no active bleeding was found in the
gastric cardia extending from GEJ to 42 cm from incisors. One malignant-appearing lymph node
was visualized in the peripancreatic region.
o Answer: Code 2 for involvement of the GE Junction/Cardia and no mention of
involvement of the stomach
EGJ is documented as involved and there is no information on stomach involvement and
o Esophagus CAP Protocol is used OR
o Esophagus Staging System is used
o If the CAP Protocol and AJCC Staging System are different, default to the AJCC Staging
System
Chapter 17: Stomach (see codes 0, 3, and 9)
Note 1: Use code 0 when only the cardia is documented as involved (no mention of EGJ)
Note 2: Use code 3 when
EGJ is documented as involved and the midpoint (epicenter) is more than 2 cm distal (below)
from the EGJ
EGJ is documented as involved and there is no information on stomach involvement AND
o Stomach CAP Protocol is used OR
o Stomach AJCC Staging System is used
o If the CAP Protocol and AJCC Staging System are different, default to the AJCC Staging
System
Note 3: Use code 9 when there is no documentation regarding EGJ involvement.

84 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description Schema ID#/Description
0 NO involvement of esophagus or gastroesophageal junction
AND epicenter at ANY DISTANCE into the proximal stomach
(including distance unknown)
00170: Stomach
2
INVOLVEMENT of esophagus or esophagogastric junction (EGJ)
AND epicenter LESS THAN OR EQUAL TO 2 cm into the proximal
stomach
OR no stated involvement of or into the stomach
00161
, 00169: Esophagus
Schemas
AND go to Schema
Discriminator 2: Histology
Discriminator for 8020/3
3
INVOLVEMENT of esophagus or esophagogastric junction (EGJ)
AND epicenter GREATER THAN 2 cm into the proximal stomach
00170: Stomach
9 UNKNOWN involvement of esophagus or gastroesophageal junction
AND epicenter at ANY DISTANCE into the proximal stomach
(including distance unknown)
00170: Stomach
<Blank> Primary site is NOT C160, Discriminator is not necessary
Return to Schema ID Table

85 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00161: Esophagus (including GE junction) Squamous (2018+)
3927: Schema Discriminator 2: Histology Discriminator for 8020/3
Item Length: 1
NAACCR Item #: 3927
XML Parent-NAACCR ID: Tumor-schemaDiscriminator2
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00161: Esophagus (including GE junction) Squamous (2018+)
00169: Esophagus (including GE junction) (excluding Squamous) (2018+)
Definition
Histology code 8020/3 is defined as “undifferentiated carcinoma.” In the AJCC 8
th
chapter for Esophagus,
this histology code is further subdivided into squamous or glandular component, which are staged
differently. A schema discriminator is necessary to distinguish between these histologies so that the
appropriate stage group table is used.
Coding Instructions and Codes
Note: A schema discriminator is used to discriminate for histology 8020/3: Undifferentiated carcinoma
to determine which AJCC Stage Group table to use.
8020/3: Undifferentiated carcinoma with squamous component (see code 1)
o Use the Squamous Cell Carcinoma AJCC Stage Group
8020/3: Undifferentiated carcinoma with glandular component (see code 2)
o Use the Adenocarcinoma AJCC Stage Group Table
8020/3: Undifferentiated carcinoma, NOS (no mention of squamous or glandular component)
(see code 3)
o Use the Squamous Cell Carcinoma AJCC Stage Group Table
Code Description Schema ID#/Description
1 Undifferentiated carcinoma with squamous component
00161: Esophagus (including GE
junction) Squamous
2 Undifferentiated carcinoma with glandular component
00169: Esophagus (including GE
junction) (excluding Squamous)
9 Undifferentiated carcinoma, NOS
00161: Esophagus (including GE
junction) Squamous
<Blank> Histology is NOT 8020, Discrminator is not necessary
Return to Schema ID Table

86 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00161: Esophagus and Esophagogastric Junction (2018+)
3829: Esophagus and EGJ Tumor Epicenter
Item Length: 1
NAACCR Item #: 3829
XML Parent-NAACCR ID: Tumor-esophagusAndEgjTumorEpicenter
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00161: Esophagus (including GE junction) Squamous (2018+)
Required for Staging: The AJCC Esophagus Staging System and EOD, for Squamous Cell Carcinomas only
Description
Esophagus and Esophagogastric Junction (EGJ), Squamous Cell (including adenosquamous), Tumor
Location refers to the position of the epicenter of the tumor in the esophagus.
Rationale
This data item is required for prognostic stage grouping for squamous and adenosquamous carcinoma in
AJCC 8
th
edition, Chapter 16 Esophagus and Esophagogastric Junction. It is a new data item for cases
diagnosed 1/1/2018 and forward.
Coding Instructions and Codes
Note 1: This data item is used for pathological staging for squamous cell carcinoma of the esophagus
and esophagogastric junction. If information is available for clinical staging, record it.
Note 2: Location is defined by the position of the epicenter of the tumor in the esophagus.
Information is most likely to be obtained from pathological exam, scopes, operative notes or CT scans.
The epicenter of the lesion is used to describe location.
Example: If the lesion was from 15-21 cm, this is a 6-cm lesion with epicenter at 18 cm. It is the
midpoint.
Note 3: Clinician or pathologist statement of epicenter being the upper, middle, or lower takes priority
over any individual results or measurements. If no statement of epicenter is provided indicating upper,
middle, or lower is provided, the following measurements may be used.
15-24 cm from incisors = upper
25-29 cm from incisors = middle
30-40/45 cm from incisors = lower
Note 4: Additional information about the epicenter may be found in Chapter 16, Esophagus and
Esophagogastric Junction, Table 16.1 and Figure 16.1.
Note 5: The ascertainment of the epicenter of the tumor is for staging purposes and is separate from the
assignment of the ICD-O-3 topography code. If you have an overlapping tumor (C158), do not recode the
topography based on the epicenter.

87 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 6: If primary site is C259 (Esophagus, NOS), code 9.
Code Description
0 U: Upper (Cervical/Proximal esophagus to lower border of azygos vein)
1 M: Middle (Lower border of azygos vein to lower border of inferior pulmonary vein)
2 L: Lower (Lower border of inferior pulmonary vein to stomach, including
gastroesophageal junction)
9 X: Esophagus, NOS
Specific location of epicenter not documented in medical record
Specific location of epicenter not assessed or unknown if assessed
Return to Schema ID Table

88 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00161: Esophagus (including GE junction) Squamous (2018+)
3855: HER2 Overall Summary
Item Length: 1
NAACCR Item #: 3855
XML Parent-NAACCR ID: Tumor-her2OverallSummary
NAACCR Alternate Name: None
Active years: 2021+
Schema(s):
00161: Esophagus (including GE junction) Squamous (2018+)
00169: Esophagus (including GE junction) (excluding Squamous) (2018+)
00170: Stomach (2018+)
Description
HER2 Overall Summary is a summary of results from HER2 testing.
Rationale
This data item is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 48 Breast. It was
previously collected as Breast, CS SSF # 15. Experts recommend that every invasive breast cancer be
tested for the presence of HER2 because anti-HER2 treatments are highly effective for these tumors.
HER2 overall summary will be collected for Esophagus and Esophagogastric Junction and Stomach for
cases diagnosed 1/1/21+ because NCCN guidelines recommend HER2 testing at time of diagnosis if
patients are documented or suspected of having metastatic disease. HER2 monoclonal antibodies may
be added to chemotherapy for patients with HER2 positive disease.
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of HER2 Overall Summary can be used to code this data item when no
other information is available.
Note 3: HER2 may be recorded for all histologies; however, it is primarily performed for
adenocarcinomas. If information is not available, code 9.
Note 4: The result of the HER2 test performed on the primary tissue is to be recorded in this data item.
Use the highest (positive versus negative) when there are multiple results
Note 5: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no HER2 results from pre-treatment specimens,
report the findings from post-treatment specimens

89 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 HER2 negative; equivocal
1 HER2 positive
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Cannot be determined (indeterminate)
HER2 Overall Summary status not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is prior to 2021
Return to Schema ID Table

90 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00169: Esophagus (including GE junction) (excluding Squamous) (2018+)
See 00161: Esophagus (including GE junction) Squamous (2018+)
3926: Schema Discriminator 1: EsophagusGEJunction (EGJ)/Stomach
3927: Schema Discriminator 2: Histology Discriminator for 8020/3
3855: HER2 Overall Summary
00170: Stomach (2018+)
See 00161: Esophagus (including GE junction) Squamous (2018+)
3926: Schema Discriminator 1: EsophagusGEJunction (EGJ)/Stomach
3855: HER2 Overall Summary
00190: Appendix (2018-2022)
See 00200: Colon and Rectum (2018+) for
3819: CEA Pretreatment Interpretation
3820: CEA Pretreatment Lab Value
09190: Appendix (2023+)
See 00200: Colon and Rectum (2018+) for
3819: CEA Pretreatment Interpretation
3820: CEA Pretreatment Lab Value
Return to Schema ID Table

91 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
09190: Appendix (2023+)
3960: Histologic Subtype
Item Length: 1
NAACCR Item #: 3960
XML Parent-NAACCR ID: histologicSubtype
NAACCR Alternate Name: None
Active years: 2023+
Schema(s):
09190: Appendix (2023+)
Definition
Histology code for appendiceal tumors (8480) is defined as “Mucinous Adenocarcinoma (in situ or
invasive).” In addition there are also low-grade appendiceal mucinous neoplasm (LAMN) and high-grade
appendiceal mucinous neoplasm (HAMN) diagnoses that are assigned the same histology.
Due to the different natures of these histologies, there is interest in tracking these different types of
tumors. With the current histology codes, a distinction cannot be made. A histology subtype data item is
needed.
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2023+.
a. For cases diagnosed 2018-2022, leave this SSDI blank
Note 2: Use the Solid Tumor Rules to determine histology prior to coding this SSDI.
Note 3: Histology 8480/2 or 8480/3 have multiple definitions that are collected in this histology. This
data item is used to further identify specific subtypes for histology code 8480/2 or 8480/3.
Examples:
1. Appendix: Disseminated peritoneal adenomucinous/low grade mucinous carcinoma peritonei.
Final diagnosis: Low grade appendiceal mucinous neoplasm
a. Code 1: This is a low grade mucinous (appendiceal) carcinoma (8480/3), which is LAMN.
The peritoneal adenomucinous/low grade mucinous carcinoma peritonei is describing
metastatic disease and not the histology
2. Appendix: Low grade (well diff) appendiceal adenocarcinoma
a. Code 0: This is an adenocarcinoma (8140/3), the low grade (well diff) is describing the
grade and not the histology
3. Appendix, appendectomy: Low grade appendiceal mucinous neoplasm (LAMN) with focal high
grade mucinous neoplasm.
a. Code 1: Based on the Solid Tumor Rules, the “focal” would be ignored and this would be
a LAMN.
4. Appendectomy: Mucinous adenocarcinoma of the appendix

92 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
a. Code 3: Mucinous adenocarcinoma is the preferred terminology for histology code
8480/3. Since there is no mention of “low grade” or “high grade”, this would not be
LAMN or HAMN
5. Appendix: Mucinous (colloid) adenocarcinoma
a. Code 3: Colloid adenocarcinoma is an alternate name for 8480/3
Code
Description
0 Histology is NOT 8480
1
Low-grade appendiceal mucinous neoplasm
LAMN
2
High-grade appendiceal mucinous neoplasm
HAMN
3
Mucinous Adenocarcinoma/carcinoma
Mucus Adenocarcinoma/carcinoma
Mucoid adenocarcinoma/carcinoma
Colloid adenocarcinoma/carcinoma
4 Other terminology coded to 8480
BLANK
NA-Diagnosis year is prior to 2023
Return to Schema ID Table

93 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3819, 3820: CEA Pretreatment Lab Value and Interpretation
Definition
A protein molecule found in many different cells of the body but associated with certain tumors and
with the developing fetus. CEA is used as a tumor marker especially for gastrointestinal cancers, as
colorectal cancer is the most frequent cause for an increased/elevated CEA. CEA is also elevated by
biliary obstruction, alcoholic hepatitis, and heavy smoking. CEA level is most frequently tested on blood
serum, but it may be tested in body fluids and/or biopsy tissue. An abnormally high CEA level prior to
tumor resection is expected to fall following successful removal of the cancer. An increasing value
indicates possible recurrence.
There are 2 data items that record information on CEA. These data items should be coded from the
same test
3819: CEA Pretreatment Interpretation
3820: CEA Pretreatment Lab Value
Coding Guidelines
Record the highest value prior to treatment in nanograms per milliliter (ng/ml) in the range 0.1 (1 ng/ml)
to 9999.9 (9999 ng/ml) in the Lab Value data item and the clinician’s interpretation of the highest value
prior to treatment, based on the reference range used by the lab in the Interpretation data item.
Examples Lab Value Code Interpretation Code
0 ng/ml 0.0 0
23.6 ng/ml 23.6 1
127.8 ng/ml 127.8 1
3567 ng/ml 3567.0 1
11,000 XXXX.1 1
Test ordered, results not in chart XXXX.7 7
Not documented in medical record
CEA test not done
Unknown if CEA test done
XXXX.9
9
Additional Information
Other names: Carcinoembryonic antigen
Source documents: clinical laboratory report, sometimes pathology or cytology report; H&P,
operative report; consultant report; discharge summary
Normal reference range:
o Nonsmoker: < 2.5 ng/ml (SI: < 2.5 mg/L) SI Conversion: 1 mg/L = 1 ng/ml.
o Smoker: < 5 ng/ml (SI: < 5 mg/L) SI Conversion: 1 mg/mL = 1 ng/L
Return to Schema ID Table

94 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3820: CEA Pretreatment Lab Value
Item Length: 6
NAACCR Item #: 3820
XML Parent-NAACCR ID: Tumor-ceaPretreatmentLabValue
NAACCR Alternate Name: CEA (Carcinoembryonic Antigen) Pretreatment Lab Value
Active years: 2018+
Schema(s):
00190: Appendix (2018-2022)
09190: Appendix (2023+)
00200: Colon and Rectum (2018+)
Description
CEA (Carcinoembryonic Antigen) Pretreatment Lab Value records the CEA value prior to treatment. CEA
is a nonspecific tumor marker that has prognostic significance for colon and rectum cancer.
Rationale
CEA (Carcinoembryonic Antigen) Pretreatment Lab Value is a Registry Data Collection Variable in AJCC.
It was previously collected as Colon and Rectum, CS SSF #3.
See 3819, 3820: CEA Pretreatment Lab Value and Interpretation for additional information.
Coding Instructions and Codes
Note 1: Physician statement of CEA (Carcinoembryonic Antigen) Pretreatment Lab Value can be used to
code this data item when no other information is available.
Note 2: Record the lab value of the highest CEA test result documented in the medical record prior to
treatment or polypectomy. The lab value may be recorded in a lab report, history and physical, or
clinical statement in the pathology report.
Note 3: CEA is a tumor marker that has value in the management of certain malignancies.
Note 4: Record to the nearest tenth in nanograms/milliliter (ng/ml) the highest CEA lab value
documented in the medical record prior to treatment or polypectomy.
Example: Code a pretreatment CEA of 7 ng/ml as 7.0.
Note 5: Record 0.1 when the lab results are stated as less than 0.1 ng/ml with no exact value.
Note 6: The same laboratory test should be used to record information in 3819: CEA Pretreatment
Interpretation.
Code Description
0.0 0.0 nanograms/milliliter (ng/ml) exactly
0.1-
9999.9
0.1-9999.9 ng/ml
(Exact value to nearest tenth in ng/ml)
XXXX.1 10,000 ng/ml or greater
XXXX.7 Test ordered, results not in chart

95 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
XXXX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XXXX.8 may result in an edit error.)
XXXX.9 Not documented in medical record
CEA (Carcinoembryonic Antigen) Pretreatment Lab Value not assessed or unknown if assessed
Return to Schema ID Table

96 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3819: CEA Pretreatment Interpretation
Item Length: 1
NAACCR Item #: 3819
XML Parent-NAACCR ID: Tumor-ceaPretreatmentInterpretation
NAACCR Alternate Name: CEA (Carcinoembryonic Antigen) Pretreatment Interpretation
Active years: 2018+
Schema(s):
00190: Appendix (2018-2022)
09190: Appendix (2023+)
00200: Colon and Rectum (2018+)
Description
CEA (Carcinoembryonic Antigen) Pretreatment Interpretation refers to the interpretation of the CEA
value prior to treatment. CEA is a glycoprotein that is produced by adenocarcinomas from all sites as
well as many squamous cell carcinomas of the lung and other sites. CEA may be measured in blood,
plasma or serum. CEA is a prognostic marker for adenocarcinomas of the appendix, colon and rectum
and is used to monitor response to treatment.
Rationale
CEA (Carcinoembryonic Antigen) is a Registry Data Collection Variable for AJCC 8. CEA
(Carcinoembryonic Antigen) Pretreatment Interpretation was previously collected as Colon and Rectum,
CS SSF #1.
See 3819, 3820: CEA Pretreatment Lab Value and Interpretation for additional information.
Coding Instructions and Codes
Note 1: Physician statement of CEA (Carcinoembryonic Antigen) Pretreatment Interpretation
can be used to code this data item when no other information is available.
Note 2: Record the interpretation of the highest CEA test result documented in the medical record prior
to treatment or a polypectomy.
Note 3: Code 3 when a CEA value was documented in the record, but there is no statement that the CEA
is positive/elevated, negative/normal, and the normal range (from which you can determine
interpretation), is not documented.
Note 4: The same laboratory test should be used to record information in 3820: CEA Pretreatment Lab
Value.
Code Description
0 CEA negative/normal; within normal limits
1 CEA positive/elevated
2 Borderline
3
Undetermined if positive or negative (normal values not available)
AND no MD interpretation
7
Test
ordered, results not in chart

97 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
8 Not applicable: Information not collected for this case
(If this data item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
CEA (Carcinoembryonic Antigen) Pretreatment Interpretation not assessed or unknown if
assessed
Return to Schema ID Table

98 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3823: Circumferential Resection Margin (CRM)
Item Length: 4
NAACCR Item #: 3823
XML Parent-NAACCR ID: Tumor-circumferentialResectionMargin
NAACCR Alternate Name: Circumferential or Radial Resection Margin (CRM)
Active years: 2018+
Schema(s):
00200: Colon and Rectum (2018+)
Description
Circumferential or Radial Resection Margin, the distance in millimeters between the leading edge of the
tumor and the surgically dissected margin as recorded on the pathology report, is a prognostic indicator
for colon and rectal cancer. This may also be referred to as the Radial Resection Margin or surgical
clearance.
Rationale
Circumferential or Radial Resection Margin is a Registry Data Collection Variable in AJCC. It was
previously collected as Colon and Rectum CS SSF #6.
Definition
The CRM, also referred to as the radial margin or the mesenteric resection margin, is the measurement
of the distance from the deepest invasion of the tumor to the margin of resection in the
retroperitoneum or mesentery. In other words, the CRM is the width of the surgical margin at the
deepest part of the tumor in an area of the large intestine or rectum without serosa (non-peritonealized
rectum below the peritoneal reflection) or only partly covered by serosa (upper rectum, posterior
aspects of ascending and descending colon).
For segments of the colon completely encased by peritoneum, the mesenteric resection margin is the
only relevant circumferential margin.
The CRM is not the same as the distance to the proximal and distal margins of the colon specimen. For
rectal cancers, the circumferential resection margin is the most important predictor of local-regional
recurrence.
Coding guidelines
Code 0.0 is for positive margins, or margin is less than 0.1 mm
Codes 0.1-99.9 is for coding the exact measurement in millimeters of the negative margin
Code XX.0 for margins described as greater than 100 mm
Code XX.1 when the margin is stated as clear, but the distance is not available
Code XX.2 when the margins cannot be assessed
Note: ONLY when the pathology reports/CAP checklist states that the margin cannot be
assessed/evaluated.
Codes XX.3-XX.6 is for when the pathology uses “at least” categories

99 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code XX.7 when there is no surgical resection of the primary site
Code XX.9 when
Not documented in the medical record
CRM is not evaluated (assessed)
Checked “Not applicable: Radial or Mesenteric Margin” on CAP Checklist
Unknown if CRM is evaluated (assessed)
Additional Information
Source documents: pathology report
For further information, refer to the Colon and Rectum cancer protocol published by the College
of American Pathologists for the AJCC Staging System Colon and Rectum
Coding Instructions and Codes
Note 1: Physician statement of Circumferential or Radial Resection Margin can be used to code this data
item when no other information is available.
Note 2: Per the AJCC Staging System Colon and Rectum, “the CRM is the distance in millimeters between
the deepest point of tumor invasion in the primary cancer and the margin of resection in the
retroperitoneum or mesentery.”
Note 3: The following guidelines were developed for the coding of surgery codes in relation to CRM.
These guidelines were confirmed by the CAP Cancer Committee.
For Colon primaries, surgery of primary site must be coded as 30-80
o If surgery of primary site is 00-29, then CRM must be coded as XX.7
For Rectal primaries, surgery of primary site must be coded as 27, 30-80
o If surgery of primary site is 00-26 or 28, then CRM must be coded as XX.7
Note 4: Tumor involvement of the circumferential resection margin or radial resection margin appears
to be a strong prognostic factor for local or systemic recurrences and survival after surgery
Note 5: The CRM may be referred to as
Circumferential radial margin
Circumferential resection margin
Mesenteric (mesocolon) (mesorectal) margin
Radial margin
Soft tissue margin
Note 6: Record in Millimeters (mm) to the nearest tenth the distance between the leading edge of the
tumor and the nearest edge of surgically dissected margin as recorded in the pathology report.
Examples
If the CRM is 2 mm, code 2.0
If the CRM is 2.78 mm, code 2.8
Note 7: If the value is recorded in Centimeters, multiply by 10 to get the value in Millimeters (mm).
Example: CRM recorded as 0.2 cm. Multiply 0.2 x 10 and record 2.0

100 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 8: If the margin is involved (positive), code 0.0. If the margin is described as less than 0.1 mm with
no more specific measurement, Code 0.0; margins of 0-1.0 mm are recorded by the pathologist as
involved.
Note 9: Code XX.2 (Margins cannot be assessed) ONLY when the pathology reports/CAP checklist states
that the margin cannot be assessed/evaluated.
Note 10: An exact measurement takes precedence over codes 0.0 and those beginning with XX.
Exact measurement takes priority even if the pathologist states the margin is positive.
Example: CRM stated as 0.3 mm in Final Diagnosis and Synoptic states: Circumferential (Radial)
Margin Interpreted as involved by invasive carcinoma (tumor less than 1mm from margin).
o Code the 0.3 mm instead of 0.0 (margin involved with tumor)
Note 11: Code XX.9 when
Checked “Not applicable: Radial or Mesenteric Margin” on CAP Checklist
Pathology report describes only distal and proximal margins, or margins, NOS
o Only specific statements about the CRM are collected in this data item
CRM not mentioned in the record
Code Description
0.0 Circumferential resection margin (CRM) positive
Margin IS involved with tumor
Described as "less than 0.1 millimeter (mm)"
0.1-
99.9
Distance of tumor from margin: 0.1- 99.9 millimeters (mm)
(Exact size to nearest tenth of millimeter)
XX.0 100 mm or greater
XX.1 Margins clear, distance from tumor not stated
Circumferential or radial resection margin negative, NOS
No residual tumor identified on specimen
XX.2 Margins cannot be assessed
XX.3 Described as “at least” 1 mm
XX.4 Described as “at least” 2 mm
XX.5 Described as “at least” 3 mm
XX.6 Described as "greater than” 3 mm
XX.7 No resection of primary site
Surgical procedure did not remove enough tissue to measure the circumferential or radial resection
margin
(Examples include: polypectomy only, endoscopic mucosal resection (EMR), excisional biopsy only,
transanal disk excision)
XX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XX.8 may result in an edit error.)
XX.9 Not documented in medical record
Non-invasive neoplasm (behavior /2)
Circumferential or radial resection margin not assessed or unknown if assessed
Return to Schema ID Table

101 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3866: KRAS
Item Length: 1
NAACCR Item #: 3866
XML Parent-NAACCR ID: Tumor-kras
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00200: Colon and Rectum (2018+)
Description
KRAS is an important signaling intermediate in the growth receptor pathway which controls cell
proliferation and survival. KRAS is a protein with production controlled by the K-ras gene. When the K-
ras gene is activated through mutation during colorectal carcinogenesis, production of KRAS
continuously stimulates cell proliferation and prevents cell deaths. Activating mutations in KRAS are an
adverse prognostic factor for colorectal carcinoma and predict a poor response to monoclonal anti-EGFR
antibody therapy in advanced colorectal carcinoma.
Rationale
KRAS is a Registry Data Collection Variable in AJCC. It was previously collected as Colon and Rectum CS
SSF #9.
Definition
KRAS is an oncogene (a gene that, when mutated or overexpressed, helps turn a normal cell into a
cancer cell). Mutations of KRAS indicate that a patient may not respond to the anti-epidermal growth
factor receptor drugs cetuximab (Erbitux) or panitumumab (Vectibix). ASCO recommends that Stage IV
colorectal patients be tested for KRAS if anti-EGFR therapy is being considered. There are two types of
KRAS genes: normal and mutated. The normal KRAS gene is also called the wild type allele; the mutated
gene may be described as abnormal or having an abnormal codon (abnormal DNA sequence).
Additional Information
Source documents: pathology report or clinical laboratory report
Other names: K-Ras, K-ras, Ki-Ras
For further information, refer to the Colon and Rectum Biomarker Reporting cancer protocol
published by the College of American Pathologists for the AJCC Staging System Colon and
Rectum
Coding Instructions and Codes
Note 1: Physician statement of KRAS can be used to code this data item when no other information is
available.
Note 2: KRAS is a gene which belongs to a class of genes known as oncogenes. When mutated,
oncogenes have the potential to cause normal cells to become cancerous. Studies suggest that KRAS
gene mutations are often present in colorectal cancer.

102 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 3: There are 4 KRAS codons that are commonly mutated in colorectal cancers. This SSDI does not
record the actual mutation, but instead records the codon or codon group that contains the mutation. If
a specific KRAS mutation is reported, its codon may be identified from the following list of common
KRAS mutations grouped by codon.
Codon 12
o Gly12Asp (GGT>GAT)
o Gly12Val (GGT>GTT)
o Gly12Cys (GGT>TGT)
o Gly12Ser (GGT>AGT)
o Gly12Ala (GGT>GCT)
o Gly12 Arg (GGT>CGT)
o Codon 12 mutation, not otherwise specified
Codon 13
o Gly13Asp (GGC>GAC)
o Gly13Arg (GGC>CGC)
o Gly13Cys (GGC>TGC)
o Gly13Ala (GGC>GCC)
o Gly13Val (GGC>GTC)
o Codon 13 mutation, not otherwise specified
Codon 61
o Gln61Leu (CAA>CTA)
o Gln61His (CAA>CAC)
o Codon 61 mutation, not otherwise specified
Codon 146
o Ala146Thr (G436A) (GCA>ACA)
o Codon 146 mutation, not otherwise specified
Note 4: KRAS analysis is commonly done for patients with metastatic disease.
Note 5: Results from nodal or metastatic tissue may be used for KRAS.
Note 6: Record the results of the KRAS from the initial workup (clinical and pathological workup).
Note 7: If KRAS is positive and there is no mention of the mutated codon, or the mutated codon is not
specified, code 4.
Note 8: Code 9 when
Insufficient amount of tissue available to perform test
No microscopic confirmation of tumor
KRAS not ordered or not done, or unknown if ordered or done
Code
Description
0 Normal
KRAS negative, KRAS wild type
Negative for (somatic) mutations, no alterations, no (somatic) mutations identified, not present,
not detected
1 Abnormal (mutated) in codon(s) 12, 13 and/or 61
2 Abnormal (mutated) in codon 146 only
3
Abnormal (mutated), but not in codon(s) 12, 13, 61, or 146

103 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
4
Abnormal (mutated), NOS, codon(s) not specified
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
KRAS not assessed or unknown if assessed
Return to Schema ID Table

104 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3890: Microsatellite Instability (MSI)
Item Length: 1
NAACCR Item #: 3890
XML Parent-NAACCR ID: Tumor-microsatelliteInstability
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00200: Colon and Rectum (2018+)
Description
Microsatellite Instability (MSI) is a form of genetic instability manifested by changes in the length of
repeated single- to six-nucleotide sequences (known as DNA microsatellite sequences). High MSI, found
in about 15% of colorectal carcinomas, is an adverse prognostic factor for colorectal carcinomas and
predicts poor response to 5-FU chemotherapy (although the addition of oxaliplatin in FOLFOX regimens
negates the adverse effects (page 266 AJCC manual)). High MSI is a hallmark of hereditary nonpolyposis
colorectal carcinoma, also known as Lynch syndrome.
Rationale
Microsatellite Instability (MSI) is a Registry Data Collection Variable in AJCC. It was previously collected
as Colon and Rectum, CS SSF #7.
Definition
Describes cancer cells that have a greater than normal number of genetic markers called microsatellites.
Microsatellites are short, repeated, sequences of DNA. Cancer cells that have large numbers of
microsatellites may have defects in the ability to correct mistakes that occur when DNA is copied in the
cell. Microsatellite instability is found most often in colorectal cancer, other types of gastrointestinal
cancer, and endometrial cancer. It may also be found in cancers of the breast, prostate, bladder, and
thyroid. Knowing whether cancer is microsatellite instability high may help plan the best treatment.
Additional Information
Other names: MSI-H
For further information, refer to the Colon and Rectum Biomarker Reporting cancer protocol
published by the College of American Pathologists for the AJCC Staging System Colon and
Rectum
Coding Instructions and Codes
Note 1: Physician statement of MSI can be used to code this data item when no other information is
available.
Note 2: The microsatellite instability (MSI) test is a genetic test performed on tumor tissue to look for
differences in length of certain non-functioning sections of DNA. The differences are caused by problems
with the genes that encode proteins that normally repair certain types of DNA damage. A high proportion

105 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
of colon cancers arising in patients with hereditary nonpolyposis colorectal cancer (HNPCC) (also known
as Lynch syndrome) have high MSI and a smaller percentage of colon cancers not associated with Lynch
syndrome have high MSI. Patients with colon cancers with high MSI may be further tested to determine
if they have HNPCC. In addition, MSI is a useful prognostic marker in that patients with high MSI colon
cancers have better response to surgery and survival.
Note 3: MSI may be recorded for all stages; however, it is primarily performed for invasive neoplasms.
For non-invasive neoplasms (behavior /2), code to 9 if no information available.
Note 4: Results from nodal or metastatic tissue may be used for Microsatellite instability.
Note 5: Testing for MSI may be done by immunology or genetic testing. Only genetic testing results will
specify whether the MSI is low or high.
MSI is looking at instability in informative markers
MSI results are recorded as
o MSS (Code 0)
o Stable (Code 0)
o Negative (Code 0)
o Low probability of MSI-H (Code 0)
o MSS/MSI-L (Code 0)
o MSI-L (Code 1)
o Unstable, high (Code 2)
o Unstable, NOS (no designation of high or low) (Code 2)
o MSI-H (Code 2)
o MSI-I (intermediate) (Code 9)
Note 6: Testing for Mismatch Repair (MMR) is usually done by immunohistochemistry (IHC).
Most common markers are MLH1, MSH2, MSH6, PMS2
MMR results are recorded as
o No loss of nuclear expression (code 0)
o Mismatch repair (MMR) intact (code 0)
o MMR proficient (pMMR or MMR-P) (code 0)
o MMR normal (code 0)
o Loss of nuclear expression (code 2)
o MMR deficient (dMMR or MMR-D) (code 2)
o MMR abnormal (code 2)
Note 7: If both tests are done and one or both are positive, code 2.
Note 8: If all tests done are negative, code 0.
Code Description
0 Microsatellite instability (MSI) stable; microsatellite stable (MSS); negative, NOS
AND/OR
Mismatch repair (MMR) intact, no loss of nuclear expression of MMR proteins
MMR proficient (pMMR or MMR-P)
1 MSI unstable low (MSI-L)
2 MSI unstable high (MSI-H)
AND/OR

106 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
MMR-D (dMMR or MMR-D), loss of nuclear expression of one or more MMR proteins, MMR
protein deficient)
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
MSI-indeterminate
MSI-equivocal
Microsatellite instability not assessed or unknown if assessed
Return to Schema ID Table

107 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3909: Perineural Invasion
Item Length: 1
NAACCR Item #: 3909
XML Parent-NAACCR ID: Tumor-perineuralInvasion
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00150: Cutaneous Carcinoma of the Head and Neck (2018+)
00200: Colon and Rectum (2018+)
00640: Skin Eyelid (2018+)
00690: Lacrimal Gland (2018+)
Description
Perineural Invasion, within or adjacent to the primary tumor, is a negative prognostic factor for
cutaneous squamous cell carcinomas of the head and neck and carcinomas of the colon and rectum,
eyelid and lacrimal gland.
Rationale
Perineural Invasion is a Registry Data Collection Variable in AJCC. It was previously collected as Colon
and Rectum CS SSF #8 and Lacrimal Gland CS SSF #4.
Definition
Perineural invasion is infiltration of nerves in the area of the lesion by tumor cells or spread of tumor
along the nerve pathway. The presence of perineural invasion has been shown in several studies to be
an indicator of poor patient prognosis. If perineural invasion is not mentioned in the pathology report,
do not assume that there is no perineural invasion.
Additional Information
Source documents: pathology report
Other names: PNI, neurotropism
For further information, refer to the Colon and Rectum cancer protocol published by the College
of American Pathologists for the AJCC Staging System Colon and Rectum
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of perineural invasion, the registrar could assume that it was negative and code
appropriately. Per the SSDI as of 2018, this assumption cannot be made. There must be a
statement that perineural invasion is not present/negative to assign “negative.” (Code 0)
Coding Instructions and Codes
Note 1: Physician statement of microscopically confirmed perineural invasion can be used to code this
data item when no other information is available.

108 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 2: Code the presence or absence of perineural invasion by the primary tumor as documented in
the pathology report.
Note 3: Information on presence of perineural invasion can be taken from either a biopsy or resection.
Absence of perineural invasion can only be taken from a surgical resection pathology report.
Note 4: Code 9 if surgical resection of the primary site is performed and there is no mention of
perineural invasion.
Code Description
0
Perineural invasion not identified/not present
Non-invasive neoplasm (behavior /2)
1 Perineural invasion identified/present
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
Pathology report does not mention perineural invasion
Cannot be determined by the pathologist
Perineural invasion not assessed or unknown if assessed
Return to Schema ID Table

109 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3934: Tumor Deposits
Item Length: 2
NAACCR Item #: 3934
XML Parent-NAACCR ID: Tumor-tumorDeposits
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00200: Colon and Rectum (2018+)
Description
A tumor deposit is defined as a discrete nodule of cancer in pericolic/perirectal fat or in adjacent
mesentery (mesocolic or rectal fat) within the lymph drainage area of the primary carcinoma, without
identifiable lymph node tissue or identifiable vascular structure.
Rationale
The presence of tumor deposits is a Registry Data Collection Variable in AJCC. It was previously
collected as Colon and Rectum CS SSF #4.
Definition
Tumor deposits are separate nodules or deposits of malignant cells in perirectal or pericolic fat without
evidence of residual lymph node tissue. If present, tumor deposits may be found within the primary
lymphatic drainage area of the tumor. They are different from direct extension from the primary tumor
and may be the result of lymphovascular invasion with extravascular extension, a totally replaced lymph
node, or discontinuous spread. Nodules of tumor outside the primary lymphatic drainage area of the
tumor are distant metastasis.
Coding guidelines
Record whether tumor deposits are present or absent.
Code 00 when the pathology report states that there are no tumor deposits
Code the number of tumor deposits reported in the pathology report. Do not count involved lymph
nodes in this field, only tumor deposits
Code X1 for 100 or more tumor deposits
Code X2 if tumor deposits are mentioned but a number is not reported
Code X9 when
o Not documented in medical record
o No surgical resection done
o Pathology report not available
o Tumor deposits not evaluated (not assessed)
o Unknown if Tumor Deposits evaluated (assessed)
Additional Information
Source documents: pathology report

110 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Other names: discontinuous extramural extension, malignant tumor foci, malignant peritumoral
deposits, satellite nodule
For further information, refer to the Colon and Rectum cancer protocol published by the College
of American Pathologists for the AJCC Staging System Colon and Rectum
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of tumor deposits, the registrar could assume there were no tumor deposits and
code none. For the SSDI, this assumption cannot be made. There must be a statement that there
are no tumor deposits to code 00.
Coding Instructions and Codes
Note 1: Physician statement of Tumor Deposits can be used to code this data item when no other
information is available.
Note 2: Tumor deposits are defined as one or more satellite peritumoral nodules in the pericolorectal
adipose tissue of a primary carcinoma without histologic evidence of residual lymph node in the
nodule.”
Tumor deposits may represent discontinuous spread, venous invasion with extravascular
spread, or a totally replaced lymph node
Note 3: Record the number of Tumor Deposits whether or not there are positive lymph nodes.
Note 4: Code X9 if surgical resection of the primary site is performed, the pathology report is available,
and tumor deposits are not mentioned.
Code Description
00 No tumor deposits
01-
99
01-99 Tumor deposits
(Exact number of Tumor Deposits)
X1 100 or more Tumor Deposits
X2 Tumor Deposits identified, number unknown
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit error.)
X9 Not documented in medical record
Cannot be determined by the pathologist
Pathology report does not mention tumor deposits
No surgical resection done
Tumor Deposits not assessed or unknown if assessed
Return to Schema ID Table

111 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3940: BRAF Mutational Analysis
Item Length: 1
NAACCR Item #: 3940
XML Parent-NAACCR ID: Tumor-brafMutationalAnalysis
Active years: 2021+
Schema(s):
00200: Colon and Rectum (2018+)
Description
The BRAF oncoprotein is involved in transmitting cell growth and proliferation signals from KRAS and
NRAS. The BRAF V600E mutation is associated with poorer prognosis and predicts lack of response to
anti-EGFR therapies.
Rationale
BRAF mutational analysis is recommended in clinical guidelines for patients with advanced colorectal
cancer as a prognostic marker and factor in determining appropriate therapy. It is a new data item for
cases diagnosed 1/1/2021+.
Definition
“BRAF V600E is a specific mutation (change) in the BRAF gene, which makes a protein that is involved in
sending signals in cells and in cell growth. This BRAF gene mutation is found in colorectal cancer. It may
increase the growth and spread of cancer cells. Checking for this BRAF mutation in tumor tissue may
help to plan cancer treatment. BRAF (V600E) kinase inhibitor RO5185426 blocks certain proteins made
by the mutated BRAF gene, which may help keep cancer cells from growing.” (NCI Dictionary of Cancer
Terms https://www.cancer.gov/publications/dictionaries/cancer-terms)
Additional Information
Source documents: pathology report or clinical laboratory report
For further information, refer to the Colon and Rectum Biomarker Reporting cancer protocol
published by the College of American Pathologists for the AJCC Staging System Colon and
Rectum
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of BRAF can be used to code this data item when no other information is
available.
Note 3: BRAF may be recorded for all stages; however, it is primarily performed for patients with
metastatic disease. If information is not available, code 9.

112 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: BRAF is a gene which belongs to a class of genes known as oncogenes. When mutated,
oncogenes have the potential to cause normal cells to become cancerous. Studies suggest that BRAF
gene mutations are often present in colorectal cancer. The most common BRAF mutations is
BRAF V600E (c.1799T>A) mutation
Note 5: The most common testing methods for BRAF are
Direct Sanger sequencing
High-resolution melting analysis
Pyrosequencing
PCR, allele-specific hybridization
Real-time PCR
Note 6: Results from nodal or metastatic tissue may be used for BRAF.
Note 7: If BRAF is positive and there is no mention of the mutated codon, or the mutated codon is not
specified, code 4.
Note 8: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no BRAF results from pre-treatment specimens,
report the findings from post-treatment specimens
Note 9: Code 9 when
Insufficient amount of tissue available to perform test
No microscopic confirmation of tumor
BRAF not ordered or not done, or unknown if ordered or done
Code
Description
0 Normal
BRAF negative, BRAF wild type
Negative for (somatic) mutations, no alterations, no (somatic) mutations identified, not present,
not detected
1 Abnormal (mutated)/detected: BRAF V600E (c.1799T>A) mutation
2 Abnormal (mutated)/detected, but not BRAF V600E (c.1799T>A) mutation
4
Abnormal (mutated), NOS
7
Te
st ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9
Not documented in medical record
BRAF not assessed or unknown if assessed
<Blank>
N/A
-
Diagnosis year is prior to 2021
Return to Schema ID Table

113 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00200: Colon and Rectum (2018+)
3941: NRAS Mutational Analysis
Item Length: 1
NAACCR Item #: 3941
XML Parent-NAACCR ID: Tumor-nrasMutationalAnalysis
NAACCR Alternate Name: None
Active years: 2021+
Schema(s):
00200: Colon and Rectum (2018+)
Description
NRAS is a signaling intermediate in the growth receptor pathway. Certain NRAS mutations predict poor
response to anti-EGFR therapy in patients with metastatic colorectal cancer.
Rationale
NRAS mutational analysis is recommended in clinical guidelines for patients with metastatic colon
cancer who are being considered for anti-EGFR therapy. It is a new data item for cases diagnosed
1/1/2021+.
Definition
KRAS (NAACCR Data Item # 3866) and NRAS are important signaling intermediates in the growth
receptor pathway, which controls cell proliferation and survival. Both KRAS and NRAS may be
constitutively activated through mutation during colorectal carcinogenesis so that they continuously
stimulate cell proliferation and prevent cell death (reference AJCC 8, pg. 266). KRAS and NRAS
mutations predict poor response to anti-EGFR therapy in patients with metastatic colon cancer. AJCC 8
estimates that KRAS may be activated in up to 40% and NRAS in about 7% of colorectal carcinomas.
Additional Information
Source documents: pathology report or clinical laboratory report
For further information, refer to the Colon and Rectum Biomarker Reporting cancer protocol
published by the College of American Pathologists for the AJCC Staging System Colon and
Rectum
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of NRAS can be used to code this data item when no other information is
available.
Note 3: NRAS may be recorded for all stages; however, it is primarily performed for patients with
metastatic disease. If information is not available, code 9.

114 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: NRAS is a gene which belongs to a class of genes known as oncogenes. When mutated,
oncogenes have the potential to cause normal cells to become cancerous. Studies suggest that NRAS
gene mutations are often present in colorectal cancer.
Note 5: There are 3 NRAS codons that are commonly mutated in colorectal cancers. This SSDI does not
record the actual mutation, but instead records the codon or codon group that contains the mutation. If
a specific NRAS mutation is reported, its codon may be identified from the following list of common
NRAS mutations grouped by codon.
Codon 12
o Gly12Asp (GGT>GAT)
o Gly12Val (GGT>GTT)
o Gly12Cys (GGT>TGT)
o Gly12Ser (GGT>AGT)
o Gly12Ala (GGT>GCT)
o Gly12Arg (GGT>CGT)
o Codon 12 mutation, not otherwise specified
Codon 13
o Codon 13 mutation, not otherwise specified
Codon 61
o Gln61Lys (CAA>AAA)
o Gln61Arg (CAA>CGA)
o Codon 61 mutation, not otherwise specified
Note 6: Results from nodal or metastatic tissue may be used for NRAS.
Note 7: If NRAS is positive and there is no mention of the mutated codon, or the mutated codon is not
specified, code 4.
Note 8: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no NRAS results from pre-treatment specimens,
report the findings from post-treatment specimens
Note 9: Code 9 when
Insufficient amount of tissue available to perform test
No microscopic confirmation of tumor
NRAS not ordered or not done, or unknown if ordered or done
Code Description
0 Normal
NRAS negative; NRAS wild type
Negative for (somatic) mutations, no alterations, no (somatic) mutations identified, not present,
not detected
1 Abnormal (mutated)/detected in codon(s) 12, 13, and/or 61
2 Abnormal (mutated)/detected, codon(s) specified but not in codon(s) 12, 13, or 61
4 Abnormal (mutated), NOS, codon(s) not specified
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)

116 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
09210: Anus (2023+)
3956: p16
Item Length: 1
NAACCR Item #: 3956
XML Parent-NAACCR ID: Tumor-p16
NAACCR Alternate Name: None
Active years: 2022+
Schema(s):
09520: Cervix (2021+)
09210: Anus (2023+)
Description
The p16 biomarker is overexpressed (produced) in response to HPV. It is therefore a surrogate marker
for HPV disease.
Rationale
Patients with HPV have a different survival or outcome so it is important to be able to distinguish this by
documenting the p16 results. Testing is performed by immunohistochemistry (IHC) which is inexpensive
and has near universal availability. It has an easily standardized interpretation. HPV testing is usually
performed through DNA testing which is more expensive and less widely available. HPV testing also has
technically more variability with the interpretation.
Definition
p16 is a tumor suppressor protein also known as cyclin-dependent kinase inhibitor 2A. The p16
biomarker is overexpressed (produced) in response to HPV. It is therefore a surrogate marker for HPV
disease.
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2023+.
For cases diagnosed 2018-2022, leave this SSDI blank
Note 2: Code 0 for p16 expression of weak intensity or limited distribution.
Note 3: This data item must be based on testing results for p16 overexpression.
A statement of a patient being HPV positive or negative is not enough to code this data item
Testing for HPV by DNA, mRNA, antibody, or other methods should not be coded in this data
item
Do not confuse p16 with HPV 16, which is a specific strain of virus
Code Description
0 p16 Negative; Nonreactive
1 p16 Positive; Diffuse, Strong reactivity
8
Not
applicable: Information not collected for this case
(If this time is required by your standard setter, use of code 8 will result in an edit error)
9 Not tested for p16; Unknown
<Blank> N/A-Diagnosis year prior to 2023
Return to Schema ID Table
117 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
HEPATOBILIARY SYSTEM

118 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3809, 3810: Alpha-Fetoprotein (AFP) Pretreatment Lab Value and Interpretation (Liver)
Definition
A protein normally produced by a fetus. Alpha fetoprotein levels are usually undetectable in the blood
of healthy adult men or women (who are not pregnant). An elevated level of alpha-fetoprotein suggests
the presence of either a primary liver cancer or germ cell tumor.
For Liver, there are 2 data items that record information on AFP. These data items should be coded
from the same test
3809: AFP Pretreatment Interpretation
3810: AFP Pretreatment Lab Value
Additional information
o Source documents: clinical laboratory report (blood serum radioimmunoassay or enzyme assay
(EIA)); sometimes in history and physical or clinical statement in pathology report
o Normal Reference Range: Adult men and non-pregnant women: 0-15 ng/ml (SI: 0-15 mg/L)
Coding guidelines
Record the highest value prior to treatment in nanograms per milliliter (ng/ml) in the range 0.1 (1 ng/ml)
to 9999.9 (9999 ng/ml) in the Lab Value data item and the clinician’s interpretation of the highest value
prior to treatment, based on the reference range used by the lab in the Interpretation data item.
Examples for AFP Pretreatment Lab Value and Interpretation
Examples Lab Value
Code
Interpretation
Code
0 ng/ml 0.0 1
23.6 ng/ml 23.6 2
127.8 ng/ml 127.8 2
3567 ng/ml 3567.0 2
11,000 XXXX.1 2
AFP test not done, or unknown if done XXXX.9 9
Return to Schema ID Table

119 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3810: AFP Pretreatment Lab Value
Item Length: 6
NAACCR Item #: 3810
XML Parent-NAACCR ID: Tumor-afpPretreatmentLabValue
NAACCR Alternate Name: AFP (Alpha Fetoprotein) Pretreatment Lab Value
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
AFP (Alpha Fetoprotein) Pretreatment Lab Value is a nonspecific serum protein that generally is elevated
in the setting of hepatocellular carcinoma (HCC). This data item pertains to the pre-treatment lab value.
Rationale
AFP (Alpha Fetoprotein) Pretreatment Lab Value is a Registry Data Collection Variable in AJCC. This data
item was previously collected for Liver, CS SSF #3.
See 3809, 3810: Alpha-Fetoprotein (AFP) Pretreatment Lab Value and Interpretation (Liver) for
additional information
Coding Instructions and Codes
Note 1: Physician statement of AFP (Alpha Fetoprotein) Pretreatment Lab Value can be used to code this
data item when no other information is available.
Note 2: Record the lab value of the highest AFP test result documented in the medical record prior to
treatment. The lab value may be recorded in a lab report, history and physical, or clinical statement in
the pathology report.
Note 3: A lab value expressed in micrograms per liter (ug/L) is equivalent to the same value expressed in
ng/ml.
Note 4: The same laboratory test should be used to record information in 3809: AFP Pretreatment
Interpretation.
Code Description
0.0 0.0 nanograms/milliliter (ng/ml); not detected
0.1-
9999.9
0.1-9999.9 ng/ml
(Exact value to nearest tenth of ng/ml)
XXXX.1 10,000.0 ng/ml or greater
XXXX.7 Test ordered, results not in chart
XXXX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XXXX.8 will result in an edit error.)
XXXX.9 Not documented in medical record
AFP (Alpha Fetoprotein) Pretreatment Lab Value not assessed or unknown if assessed
Return to Schema ID Table

120 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3809: AFP Pretreatment Interpretation
Item Length: 1
NAACCR Item #: 3809
XML Parent-NAACCR ID: Tumor-afpPretreatmentInterpretation
NAACCR Alternate Name: AFP (Alpha Fetoprotein) Pretreatment Interpretation
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
AFP (Alpha Fetoprotein) Pretreatment Interpretation, a nonspecific serum protein that generally is
elevated in the setting of hepatocellular carcinoma (HCC), is a prognostic factor for liver cancer.
Rationale
AFP (Alpha Fetoprotein) Pretreatment Interpretation is a Registry Data Collection Variable in AJCC. This
data item was previously collected for Liver, CS SSF #1.
See 3809, 3810: Alpha-Fetoprotein (AFP) Pretreatment Lab Value and Interpretation (Liver) for
additional information
Coding Instructions and Codes
Note 1: Physician statement of AFP (Alpha Fetoprotein) Pretreatment Interpretation can be used to
code this data item when no other information is available.
Note 2: Record the interpretation of the highest AFP test result documented in the medical record prior
to treatment.
Note 3: The same laboratory test should be used to record information in 3810: AFP Pretreatment Lab
Value.
Code Description
0 Negative/normal; within normal limits
1 Positive/elevated
2 Borderline; undetermined if positive or negative
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
AFP pretreatment interpretation not assessed or unknown if assessed
Return to Schema ID Table

121 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score
The Model for End-stage Liver Disease (MELD) score is used to assess the severity of chronic liver
disease, and its original purpose was to help prioritize patients for liver transplant by estimating their
risk of dying while waiting for transplant. There are several data items that are defined to record the
MELD score.
3813: Bilirubin Pretreatment Total Lab Value
3814: Bilirubin Pretreatment Unit of Measure
3824: Creatinine Pretreatment Lab Value
3825: Creatinine Pretreatment Unit of Measure
3860: International Normalized Ratio
Bilirubin Pretreatment Lab Value and Unit of Measure
Bilirubin is produced from the breakdown of hemoglobin (the protein that binds oxygen) in red blood
cells. The liver processes bilirubin by excreting it through bile into the intestine. If the liver is damaged,
there will be too much bilirubin in the blood, and this can produce jaundice. Elevated bilirubin levels can
indicate liver or blood disorders or blockage of bile ducts. Do not code individual conjugate, direct,
unconjugated, indirect, or delta values or bilirubin in urine.
There are two methods of describing bilirubin levels in the blood, weight in grams (milligrams per
deciliter), usually used in the US, and molecular count in moles (micromoles per liter) in Canada.
Conversion: 1 mg/dL = 17.1 mol/L. Code the unit of measure used by the facility laboratory.
Coding guidelines
Record the highest value prior to treatment in the range 0.1 (1 ng/ml) to 999.9 in the Lab Value data
item and the unit of measure for measuring the lab value in the Unit of Measure data item.
Examples Lab Value
Code
Unit of Measure
Code
0 ng/ml 0.0 1
23.6 umol/L 23.6 2
127.8 ng/mL 127.8 1
1567 umol/mL XXX.1 2
638.4 638.4 9
Test ordered, results not in chart XXXX.7 7
Not documented in medical record
Bilirubin test not done
Unknown if Bilirubin test done
XXXX.9 9
Additional Information
o Source documents: clinical laboratory report (blood serum); value may be part of a metabolic or
liver function panel

122 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Other names: TBIL. Total bilirubin is a combination of direct (conjugated), indirect
(unconjugated), and delta (conjugated bilirubin bound to albumin) bilirubin levels
o Normal Reference Range: 0.3-1.5 mg/dL (5-20.5 mol/L). The normal range may vary slightly
from lab to lab.
o For Liver, there are 2 data items that record information on Bilirubin. These data items
should be coded from the same test
o Bilirubin Pretreatment Total Lab Value [NAACCR Data Item #3813]
o Bilirubin Pretreatment Unit of Measure [NAACCR Data item #3814]
Creatinine Pretreatment Lab Value and Unit of Measure
Creatinine concentration in blood is a marker of renal function. Elevated levels are associated with
severe liver disease. Creatinine can be measured in blood serum or urine, but these data items apply to
blood levels only. Do not code urine creatinine or creatinine clearance in this field.
There are two methods of describing creatinine levels in the blood, weight in grams (milligrams per
deciliter), usually used in the US, and molecular count in moles (micromoles per liter) in Canada.
Conversion: 1 mg/dL = 17.1 mol/L. Code the unit of measure used by the facility laboratory.
Coding guidelines
Record the highest value prior to treatment in the range 0.1 (1 ng/ml) to 999.9 in the Lab Value data
item and the unit of measure for measuring the lab value in the Unit of Measure data item.
Examples Lab Value Code Unit of Measure Code
0 ng/ml 0.0 1
0.7 umol/L 0.7 2
25.4 ng/ml 25.4 1
127.6 umol/L XX.1 2
98.3 98.3 9
Test ordered, results no in chart XXXX.7 7
Not documented in medical record
Creatinine test not done
Unknown if Creatinine test done
XXXX.9 9
Additional Information
Source documents: clinical laboratory report (blood serum); value may be part of a metabolic
panel
Other names: Serum creatinine, plasma creatinine (PCr), blood creatinine, Creat, Cre.
o Do not confuse with creatinine clearance or creatine; these are unrelated tests. Do not
code urine creatinine or creatinine clearance.
Normal Reference Range
o Women: 0.5-1.0 mg/dL (45-90 mmol/L)
o Men: 0.7-1.2 mg/dL (60-110 mmol/L). Male values are usually higher due to greater
muscle mass.
o Normal value ranges may vary slightly among different laboratories.

123 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
International Normalized Ratio (Prothrombin Time)
The prothrombin time is a measure of how quickly the blood clots, which may also indicate liver disease.
The international normalized ratio (INR) is a calculation of the patient’s prothrombin time divided by the
normal mean prothrombin time for the particular thromboplastin reagent used and is expressed as a
decimal number. An elevated level indicates the blood is too “thin” and does not clot properly,
increasing the risk of bleeding. A value under 1.0 increases the risk of blood clots.
Coding guidelines
Code the highest INR value in the blood prior to treatment in the range 0.1 to 9.9.
Code X.1 for an INR of 10.0 or greater.
Code X.7 if the test was ordered and the results are not in the medical record.
Code X.9 when
o there is no information in the medical record about the INR or prothrombin time
o the test is not done or it’s unknown if the test was done
Additional Information
Source documents: clinical laboratory report (blood serum); value may be part of a metabolic or
liver function panel; outpatient or ambulatory blood test (finger stick) reported in patient
history
Other names: INR
Normal ranges: For a healthy person is 0.9-1.3. A high INR level such as INR=5 indicates that
there is a high chance of bleeding. A low level such as INR = 0.5 indicates a high chance of
abnormal clotting. Normal values may vary from lab to lab. Record the highest INR value prior to
treatment.

124 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3813: Bilirubin Pretreatment Total Lab Value
Item Length: 5
NAACCR Item #: 3813
XML Parent-NAACCR ID: Tumor-bilirubinPretxTotalLabValue
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
Bilirubin Pretreatment Total Lab Value records the bilirubin value prior to treatment. Bilirubin level is an
indicator of how effectively the liver excretes bile and is required to calculate the Model for End-Stage
Liver Disease (MELD) score used to assign priority for liver transplant.
Rationale
Bilirubin Pretreatment Total Lab Value is a Registry Data Collection Variable in AJCC. This data item was
previously collected as Liver, CS SSF #6.
See 3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score for additional
information
Coding Instructions and Codes
Note 1: Physician statement of Bilirubin Pretreatment Total Lab Value can be used to code this data item
when no other information is available.
Note 2: Record the lab value of the highest Bilirubin Total test results documented in the medical record
prior to treatment. The lab value may be recorded in a lab report, history and physical, or clinical
statement in the pathology report.
Note 3: Assay of Bilirubin Pretreatment Total Lab Value includes conjugated (direct) and unconjugated
(indirect) bilirubin and total bilirubin values. Record the total bilirubin value for this data item.
Note 4: Record to the nearest tenth of mg/dL or umol/L the highest total bilirubin value prior to
treatment.
Note 5: The Model for End-Stage Liver Disease (MELD) is a numerical scale used to determine how
urgently a patient with liver disease needs a liver transplant. Results from three routine lab tests are
used to calculate the MELD score. Bilirubin, one of the tests, measures how effectively the liver excretes
bile.
Note 6: The same laboratory test should be used to record information in 3814: Bilirubin Pretreatment
Unit of Measure.

125 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0.0 0.0 milligram/deciliter (mg/dL)
0.0 micromole/liter (umol/L)
0.1-
999.9
0.1-999.9 milligram/deciliter (mg/dL)
0.1-999.9 micromole/liter (umol/L)
XXX.1 1000 milligram/deciliter (mg/dL) or greater
1000 micromole/liter (umol/L) or greater
XXX.7 Test ordered, results not in chart
XXX.8
Not applicable:
Information not collected
for this case
(If this item is required by your standard setter, use of code XXX.8 will result in an edit error.)
XXX.9
Not documented in medical record
Bilirubin Pretreatment Total Lab Value not assessed or unknown if assessed
Return to Schema ID Table

126 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3814: Bilirubin Pretreatment Unit of Measure
Item Length: 1
NAACCR Item #: 3814
XML Parent-NAACCR ID: Tumor-bilirubinPretxUnitOfMeasure
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
Bilirubin Pretreatment Unit of Measure identifies the unit of measure for the bilirubin value measured
prior to treatment. Bilirubin is commonly measured in units of Milligrams/deciliter (mg/dL) in the United
States and Micromoles/liter (umol/L) in Canada and Europe.
Rationale
Bilirubin Pretreatment is a Registry Data Collection Variable in AJCC. Bilirubin Pretreatment Unit of
Measure is needed to identify the unit in which bilirubin is measured and was previously collected as
Liver, CS SSF #7.
See 3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score for additional
information
Coding Instructions and Codes
Note 1: Physician statement of Bilirubin Pretreatment Unit of Measure can be used to code this data
item when no other information is available.
Note 2: There are two main methods of describing concentrations: by weight, and by molecular count.
Weights are recorded in grams, and molecular counts are recorded in moles.
Milligrams/deciliter (mg/dL) is the unit of measure commonly used in the United States.
Micromoles/liter (umol/L) is the designated Systeme International (SI) unit of measure
commonly used in Canada and Europe.
1 mg/dL of bilirubin is 17.1 umol/L.
Note 3: The same laboratory test should be used to record information in 3813: Bilirubin Pretreatment
Total Lab Value.
Code
Description
1 Milligrams per deciliter (mg/dL)
2 Micromoles/liter (umol/L)
7 Test ordered, results not in chart
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Bilirubin unit of measure not assessed or unknown if assessed
Return to Schema ID Table

127 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3824: Creatinine Pretreatment Lab Value
Item Length: 4
NAACCR Item #: 3824
XML Parent-NAACCR ID: Tumor-creatininePretreatmentLabValue
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
Creatinine Pretreatment Lab Value, an indicator of kidney function, is required to calculate the Model
for End-Stage Liver Disease (MELD) score, which is used to assign priority for liver transplant.
Rationale
Creatinine Pretreatment Lab Value is a Registry Data Collection Variable in AJCC. This data item was
previously collected for Liver, CS SSF #4.
See 3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score for additional
information
Coding Instructions and Code
Note 1: Physician statement of Creatinine Pretreatment Lab Value can be used to code this data item
when no other information is available.
Note 2: Record the lab value of the highest Creatinine test result documented in the medical record
prior to treatment. The lab value may be recorded in a lab report, history and physical, or clinical
statement in the pathology report.
Note 3: Record the blood or serum creatinine value for this data item. Do not use urine results to code
this data item.
Note 4: The Model for End-Stage Liver Disease (MELD) is a numerical scale used to determine how
urgently a patient with liver disease needs a liver transplant within the next three months. Results from
three routine lab tests are used to calculate the MELD score. Creatinine, one of the tests, measures
kidney function; impaired kidney function is often associated with severe liver disease.
Note 5: The same laboratory test should be used to record information in 3825: Creatinine Pretreatment
Unit of Measure.
Code Description
0.0
0.0 milligram/deciliter (mg/dl)
0.0 micromole/liter (umol/L)
0.1-99.9 0.1-99.9 milligram/deciliter (mg/dl)
0.1-99.9 micromole/liter (umol/L)
(Exact value to nearest tenth of mg/dl or umol/L)

128 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
XX.1 100 mg/dl or greater
100 umol/L or greater
XX.7 Test ordered, results not in chart
XX.8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code XX.8 will result in an edit error.)
XX.9
Not documented in medical record
Creatinine Pretreatment Lab Value not assessed or unknown if assessed
Return to Schema ID Table

129 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3825: Creatinine Pretreatment Unit of Measure
Item Length: 1
NAACCR Item #: 3825
XML Parent-NAACCR ID: Tumor-creatininePretxUnitOfMeasure
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
Creatinine Pretreatment Unit of Measure identifies the unit of measure for the creatinine value
measured in blood or serum prior to treatment. Creatinine is commonly measured in units of
Milligrams/deciliter (mg/dL) in the United States and Micromoles/liter (umol/L) in Canada and Europe.
Rationale
Creatinine Pretreatment is a Registry Data Collection Variable in AJCC. Creatinine Pretreatment Unit of
Measure is needed to identify the unit in which creatinine is measured and was previously collected as
Liver, CS SSF #5.
See 3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score for additional
information
Coding Instructions and Codes
Note 1: Physician statement of Creatinine Pretreatment Unit of Measure can be used to code this data
item when no other information is available.
Note 2: There are two main methods of describing concentrations: by weight, and by molecular count.
Weights are recorded in grams, and molecular counts are recorded in moles.
Milligrams/deciliter (mg/dL) is the unit of measure commonly used in the United States
Micromoles/liter (umol/L) is the designated Systeme International (SI) unit of measure
commonly used in Canada and Europe.
1 mg/dL of creatinine is 88.4 umol/L.
Note 3: The same laboratory test should be used to record information in 3824: Creatinine Pretreatment
Lab Value.
Code
Description
1 Milligrams/deciliter (mg/dL)
2 Micromoles/liter (umol/L)
7 Test ordered, results not in chart
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Creatinine unit of measure not assessed or unknown if assessed
Return to Schema ID Table

130 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3860: International Normalized Ratio
Item Length: 3
NAACCR Item #: 3860
XML Parent-NAACCR ID: Tumor-iNRProthrombinTime
NAACCR Alternate Name: INR (International Normalized Ratio for Prothrombin Time)
Active years: 2018+
Schema(s):
00220: Liver (2018+)
Description
International Normalized Ratio for Prothrombin Time (INR), an indicator of the liver’s ability to make
clotting factors, is required to calculate the Model for End-Stage Liver Disease (MELD) score, is used to
assign priority for liver transplant.
Rationale
International Normalized Ratio for Prothrombin Time (INR) is a Registry Data Collection Variable in AJCC.
This data item was previously collected for Liver, CS SSF #8.
See 3813-3814, 3824-3825, 3860: Model for End-Stage Liver Disease (MELD) Score for additional
information
Coding Instructions and Codes
Note 1: Physician statement of the International Normalized Ratio for Prothrombin Time (INR) can be
used to code this data item when no other information is available.
Note 2: Record the value of the highest INR test results documented in the medical record prior to
treatment. The value may be recorded in a lab report, history and physical, or clinical statement in the
pathology report.
Note 3: The Model for End-Stage Liver Disease (MELD) is a numerical scale used to determine how
urgently a patient with liver disease needs a liver transplant. Results from three routine lab tests are
used to calculate the MELD score. International normalized ratio for prothrombin time (INR), one of the
tests, measures the liver's ability to make blood clotting factors.
Code Description
0.0 0.0
0.1
0.1 or less
0.2
-
9.9
0.2
-
9.9
(Exact ratio to nearest tenth)
X.1
10 or greater
X.7 Test ordered, results not in chart
X.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X.8 may result in an edit error.)
X.9 Not documented in medical record
INR (International Normalized Ratio for Prothrombin Time) not assessed or unknown if assessed
Return to Schema ID Table

131 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00220: Liver (2018+)
3835: Fibrosis Score
Item Length: 1
NAACCR Item #: 3835
XML Parent-NAACCR ID: Tumor-fibrosisScore
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00220: Liver (2018+)
00230: Bile Ducts Intrahepatic (2018+)
Description
Fibrosis Score (Ishak Score), the degree of fibrosis of the liver based on pathological examination, is a
prognostic factor for liver cancer.
Rationale
Fibrosis Score is a Registry Data Collection Variable in AJCC. This data item was previously collected for
Liver, CS SSF #2.
Definition
The Fibrosis Score is based on degree of parenchymal fibrosis or cirrhosis of the nontumorous liver as
defined in the surgical pathology report. Multiple fibrosis scoring systems have been described for use in
pathological evaluation of liver disease.
Ishak system uses a scale of 0-6 with 6 indicating cirrhosis.
o Recommended by AJCC and CAP
Batts-Ludwig system uses a score of 0-4, with a score of 3 defined as fibrous septa with
architectural distortion but no obvious cirrhosis, and a score of 4 defined as cirrhosis
o Used most commonly by US pathologists
METAVIR uses scores of F0-F4
o Used mostly in Europe
Additional Information
Source documents: pathology report (biopsy or FNA path report), surgical resection
Other names: Nontumoral hepatic parenchymal fibrosis/cirrhosis (Intrahepatic Bile Duct
Tumors)
Coding Instructions and Codes
Note 1: Physician statement of fibrosis score can be used to code this data item when no other
information is available. However, code 7 when the physician statement of fibrosis score is not based on
histologic examination of the liver.
Note 2: FIB-4 is NOT a pathological fibrosis score of 4. It is a scoring method using the patient’s age and
relevant lab values to calculate a score. The medical record may show something like “FIB-4 = 3.52.” Do
not code FIB-4 values in this data item.

132 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 3: AJCC classifies Ishak fibrosis scores 0-4 (none to moderate fibrosis) as F0, and Ishak fibrosis
scores 5-6 (cirrhosis/severe fibrosis) as F1. This is not the same as METAVIR score F0 or F1.
Note 4: Record the results based on information collected during the initial work-up through the first
course surgery, in the absence of neoadjuvant treatment. If multiple histologic assessments of the liver
(biopsies or resections) are taken and have conflicting scores, record the highest score.
Information collected after the start of neoadjuvant treatment or primary systemic or radiation
therapy may not be used to code this data item.
Note 5: To use codes 0 and 1, you must have a histological (microscopic) confirmation of
fibrosis/cirrhosis. Code the absence (code 0) or presence (code 1) of fibrosis as documented in the
pathology report.
Note 6: Use code 7 if there is a clinical diagnosis (no microscopic confirmation) of severe fibrosis or
cirrhosis.
Note 7: If no score is mentioned, descriptive terms may be used to assign codes 0 and 1 – see specific
terms in the table below.
Note 8: If a fibrosis score is stated but the scoring system is not recorded, consult with the physician. If
no further information is available, code 9.
Code Description
0
Any of the following histologically confirmed
No to moderate fibrosis
Ishak fibrosis score 0-4
METAVIR score F0-F3
Batt-Ludwig score 0-3
1
Any of the following histologically confirmed
Advanced/severe fibrosis
Developing cirrhosis
Incomplete cirrhosis
Transition to cirrhosis
Cirrhosis, probably or definite
Cirrhosis, NOS
Ishak fibrosis score 5-6
METAVIR score F4
Batt-Ludwig score 4
7
Clinical statement of advanced/severe fibrosis or cirrhosis, AND
Not histologically confirmed or unknown if histologically confirmed
8
Not applicable: Information not collected for this
case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Stated in medical record that patient does not have advanced cirrhosis/advanced fibrosis, not
histologically confirmed or unknown if histologically confirmed
Fibrosis score stated but cannot be assigned to codes 0 or 1
Fibrosis score stated but scoring system not recorded
Fibrosis Score not assessed or unknown if assessed
Return to Schema ID Table

134 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00230: Bile Ducts Intrahepatic (2018+)
3917: Primary Sclerosing Cholangitis
Item Length: 1
NAACCR Item #: 3917
XML Parent-NAACCR ID: Tumor-primarySclerosingCholangitis
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00230: Bile Ducts Intrahepatic (2018+)
00250: Bile Ducts Perihilar (2018+)
Description
Primary sclerosing cholangitis denotes a chronic autoimmune inflammation of the bile ducts that leads
to scar formation and narrowing of the ducts over time. It is a prognostic factor for intrahepatic bile duct
cancer.
Rationale
Primary Sclerosing Cholangitis is a Registry Data Collection Variable in AJCC. This data item was
previously collected for Intrahepatic Bile Duct, SSF #11.
Definition
Primary sclerosing cholangitis is an idiopathic liver disease characterized by inflammation and fibrosis of
the entire biliary tree. The chronic inflammation and injury to ducts may lead to cirrhosis and predispose
to cholangiocarcinoma at any site in the biliary tree. Patients with primary sclerosing cholangitis are
advised to receive neoadjuvant chemoradiation and liver transplantation.
Coding guidelines
Record whether primary sclerosing is absent or present
Code 0 when there is a statement in the pathology report that primary sclerosing cholangitis is
not present
Code 1 when the pathology report states that primary sclerosing cholangitis is present
Code 9 when
o No information in the medical record
o Pathology report is not available
o Primary Sclerosing Cholangitis is not evaluated (not assessed)
o Unknown if Primary Sclerosing Cholangitis is evaluated (assessed)
Additional Information
Source documents: patient history, pathology report, imaging reports
Other names: PSC, fibrosing cholangitis, chronic obliterative cholangitis, sclerosing cholangitis
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of primary sclerosing cholangitis, the registrar could assume that it was not

135 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
present and code appropriately. For the SSDI, this assumption cannot be made. There must be a
statement that primary sclerosing cholangitis is not present to code 0.
Coding Instructions and Codes
Note 1: Physician statement of Primary Sclerosing Cholangitis (PSC) can be used to code this data item
when no other information is available.
Note 2: PSC is an idiopathic liver disease characterized by inflammation and fibrosis of the entire biliary
tree. The chronic inflammation and injury to ducts may lead to cirrhosis and predispose to
cholangiocarcinoma at any site in the biliary tree.
Note 3: Code stated diagnosis of PSC either clinically or pathologically as documented in the medical
record. This may be by history.
Note 4: Code 9 if there is no mention of primary sclerosing cholangitis (PSC).
Code Description
0 PSC not identified/not present
1 PSC present
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
PSC not assessed or unknown if assessed
Return to Schema ID Table

136 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00230: Bile Ducts Intrahepatic (2018+)
3935: Tumor Growth Pattern
Item Length: 1
NAACCR Item #: 3935
XML Parent-NAACCR ID: Tumor-tumorGrowthPattern
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00230: Bile Ducts Intrahepatic (2018+)
Description
Tumor Growth Pattern refers to the growth pattern of intrahepatic cholangiocarcinoma.
Rationale
Tumor Growth Pattern is a Registry Data Collection Variable in AJCC. This data item was previously
collected for Intrahepatic Bile Duct, SSF #10.
Definition
There are two types of growth patterns for intrahepatic bile duct carcinomas.
Mass-forming (60% of intrahepatic bile duct cases), which grows outward (radially) from the
duct and invades the liver parenchyma in a well-defined mass.
Periductal infiltrating type (20%): spreads along the duct in a diffuse manner that may be
associated with poorer prognosis.
Coding guidelines
Record the specific type of tumor growth pattern.
Code 1 when a radiology, surgery, or pathology report describes the tumor as mass-forming only
Code 2 when a radiology, surgery, or pathology report describes the tumor as periductal
infiltrating only
Code 3 when a radiology, surgery, or pathology reports mentions both mixed mass forming and
periductal infiltrating
Code 9
Not documented in the medical record
Tumor growth pattern not evaluated (assessed)
Unknown if Tumor Growth Pattern evaluated (assessed)
Additional Information
Source documents: radiology, surgery, or pathology report
Coding Instructions and Codes
Note 1: Physician statement of tumor growth pattern can be used to code this data item when no other
information is available.

137 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 2: Cholangiocarcinoma may be classified by growth pattern. The tumor growth patterns of
intrahepatic cholangiocarcinoma include the mass forming type, the periductal infiltrating type, and a
mixed type. The periductal infiltrating type of cholangiocarcinoma demonstrates a diffuse longitudinal
growth pattern along the bile duct. Limited analyses suggest that the diffuse periductal infiltrating type
is associated with a poor prognosis.
Note 3: Record the presence or absence of an infiltrating periductal component. This information may
be obtained from radiology, surgery, or pathology reports.
Code Description
1 Mass-forming
2 Periductal infiltrating
3 Mixed mass-forming and periductal infiltrating
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
Radiology and/or pathology report does not mention tumor growth pattern
Cannot be determined by the pathologist
Tumor growth pattern not assessed or unknown if assessed
Return to Schema ID Table

138 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00242: Cystic Duct (2018+)
3926: Schema Discriminator 1: BileDuctsDistal/BileDuctsPerihilar/CysticDuct
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00242: Cystic Duct (2018+)
00250: Bile Ducts Perihilar (2018+)
00260: Bile Duct Distal (2018+)
Definition
Cystic duct, distal bile ducts, and perihilar bile ducts all have the same ICD-O topography code (C240).
However, for purposes of stage grouping in the AJCC 8
th
edition, they each have different chapters for
stage. A schema discriminator is necessary to distinguish between these primary sites so that the
appropriate chapter/schema is used.
Coding Instructions and Codes
Note: A schema discriminator is used to discriminate for primary site C240 (extrahepatic bile ducts) for
the subsite in which the tumor arose.
00242: Cystic Duct (see code 3)
Per the AJCC Gallbladder Staging System, the gallbladder tapers into the cystic duct
00250: Bile Ducts Perihilar (see codes 1, 5, 6, 9)
Per the AJCC Perihilar Bile Ducts Staging System, perihilar (or proximal) cholangiocarcinomas
involve the main biliary confluence of the right and left hepatic ducts and comprise 50%-70% of
all cases of bile duct carcinoma
00260: Bile Ducts Distal (see codes 4, 7)
Per the AJCC Distal Bile Duct Staging System, these tumors have their center located between
the confluence of the cystic duct and common hepatic duct and the Ampulla of Vater (excluding
ampullary carcinomas)
Code Description Schema ID#/Description
1 Perihilar bile duct(s)
Proximal extrahepatic bile duct(s)
Hepatic duct(s)
00250: Bile Ducts Perihilar
3 Cystic bile duct; cystic duct 00242: Cystic Duct
4
Distal bile duct
Common bile duct
Common duct, NOS
00260: Bile Duct Distal
5
Diffuse involvement
More than one subsite involved, subsite of origin not stated
00250: Bile
Ducts Perihilar
6
Stated as middle extrahepatic bile duct
AND treated with combined hepatic and hilar resection
00250: Bile Ducts Perihilar

140 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00250: Bile Ducts Perhilar (2018+)
See 00242: Cystic Duct (2018+)
3926: Schema Discriminator 1: BileDuctsDistal/BileDuctsPerihilar/CysticDuct
See 00230: Bile Ducts Intrahepatic
3917: Primary Sclerosing Cholangitis
00260: Bile Duct Distal (2018+)
See 00242: Cystic Duct (2018+)
3926: Schema Discriminator 1: BileDuctsDistal/BileDuctsPerihilar/CysticDuct
Return to Schema ID Table

141 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00280: Pancreas (2018+)
3942: CA 19-9 PreTx Lab Value
Item Length: 6
NAACCR Item #: 3942
XML Parent-NAACCR ID: Tumor-ca199PretxLabValue
Alternate Name: Carbohydrate Antigen 19-9 Pretreatment Lab Value
Active years: 2021+
Schema(s):
00280: Pancreas (2018+)
Description
Carbohydrate Antigen (CA) 19-9 Pretreatment Lab Value records the CA 19-9 value prior to treatment.
CA 19-9 is a tumor marker that has prognostic significance for pancreatic cancer.
Rationale
CA 19-9 Pretreatment Lab Value is a strong predictor of resectability in the absence of metastatic
disease. It is a new data item for cases diagnosed 1/1/2021+.
Definition
CA 19-9 is a sialylated Lewis A blood group antigen that is commonly expressed and shed in pancreatic
and hepatobiliary disease and in many malignancies, thus is not tumor specific. Preoperative CA 19-9
levels in pancreatic cancer patients correlate both with AJCC staging and resectability [NCCN Guidelines
Version 3.2019 Pancreatic Adenocarcinoma].
Additional Information
Source documents: clinical laboratory report
Other names: Carbohydrate Antigen 19-9, Cancer Antigen-GI, CA-GI, Cancer Antigen 19-9
https://labtestsonline.org/tests/cancer-antigen-19-9#
Coding Instructions and Code
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of CA 19-9 (Carbohydrate Antigen 19-9) Pretreatment Lab Value can be
used to code this data item when no other information is available.
Note 3: Record the lab value of the highest CA 19-9 test result documented in the medical record prior
to treatment. The lab value may be recorded in a lab report, history and physical, or clinical statement
in the pathology report.
Note 4: A known lab value takes priority over codes XXXX.2 and XXXX.3
The lab value takes priority even if the physician documents the interpretation
Example: Patient noted to have a CA 19-9 of 3,219. Physician notes that the value is elevated.

142 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Code 3219.0 instead of XXXX.3 (elevated)
Note 5: CA 19-9 is a tumor marker that has value in the management of certain malignancies.
Note 6: Record to the nearest tenth in Units/milliliter (U/ml), the highest CA 19-9 lab value documented
in the medical record prior to treatment.
Example 1: Code a pretreatment CA 19-9 of 7 U/ml as 7.0
Example 2: Code a pretreatment CA 19-9 of 1672.3 U/ml as 1672.3
Note 7: Record 0.1 when the lab results are stated as less than 0.1 U/ml with no exact value.
Code Description
0.0 0.0 Units/milliliter (U/ml) exactly
0.1-
9999.9
0.1-9999.9 U/ml
(Exact value to nearest tenth in U/ml)
XXXX.1 10,000 U/ml or greater
XXXX.2 Lab value not available, physician states CA 19-9 is negative/normal
XXXX.3 Lab value not available, physician states CA 19-9 is positive/elevated/high
XXXX.7 Test ordered, results not in chart
XXXX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XXXX.8 may result in an edit error.)
XXXX.9 Not documented in medical record
CA (Carbohydrate Antigen) 19-9 Pretreatment Lab Value not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is prior to 2021
Return to Schema ID Table

143 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00290: NET Stomach (2018+)
3863: Ki-67
Item Length: 5
NAACCR Item #: 3863
XML Parent-NAACCR ID: Tumor-ki67
Active years: 2021+
Schema(s):
00290: NET Stomach (2018+)
00301: NET Duodenum (2018+)
00302: NET Ampulla of Vater (2018+)
00310: NET Jejunum and Ileum (2018+)
00320: NET Appendix (2018+)
00330: NET Colon and Rectum (2018+)
00340: NET Pancreas (2018+)
Description
Ki-67 (MIB-1) (Proliferative Index) is a marker of cell proliferation. A high value indicates a tumor that is
proliferating more rapidly. Codes and coding instructions for this data item are site-specific.
Rationale
Ki-67 (MIB-1) (Proliferative Index) is a Registry Data Collection Variable in AJCC. It was a new data item
for breast cases diagnosed 1/1/2018+. It will apply to neuroendocrine tumors (NET) of the
gastrointestinal tract (AJCC Chapters 29 – 34) for cases diagnosed 1/1/2021+. High Ki-67 is an adverse
prognostic factor and Ki-67 is a component of grade for these tumors. NCCN guidelines recommend
that tumor differentiation, mitotic rate and Ki-67 should be recorded in the pathology report for these
tumors.
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of Ki-67 (MIB-1), also referred to as the “Proliferative Index” can be used to
code this data item.
Note 3: Ki-67 is a marker of cell proliferation. A high value indicates a tumor that is proliferating more
rapidly.
Note 4: Results from nodal or metastatic tissue may not be used.
If the only information you have have is a Ki-67 from a metastatic site, code to XXX.9
Note 5: Ki-67 results are reported as the percentage cell nuclei that stain positive. As of early 2017,
there are no established standards for interpretation of results or for cutoffs for positive and negative.
Example 1: Ki-67 reported as 14%. Code 14.0
Example 2: Ki-67 reported as 8.6%. Code 8.6
Note 6: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.

144 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
If neoadjuvant therapy is given and there are no Ki-67 results from pre-treatment specimens,
report the findings from post-treatment specimens
Note 7: A specific value (0.0-100.0) takes priority over XXX.4, XXX.5 or XXX.6. Code the exact percentage
when provided. When the exact percentage is not given, including ranges or terms such as “less than” or
“greater than” use the range value codes XXX.4, XXX.5, XXX.6.
XXX.4, XXX.5 and XXX.6 were added since they are listed on the CAP protocol
o Example 1: Ki-67 stated as less than 1%. Code XXX.4
o Example 2: Ki-67 stated as 5%-10%. Code XXX.5
o Example 3: Ki-67 stated as greater than 4%. Code XXX.5
o Example 4: Ki-67 stated as greater than 30%. Code XXX.6
Code Description
0.0-100.0 0.0 to 100.0 percent positive: enter percent positive
XXX.4 Ki-67 stated as less than 3%
XXX.5 Ki-67 stated as 3%-20%
XXX.6 Ki-67 stated as greater than 20%
XXX.7 Test done; actual percentage not stated
XXX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XXX.8 will result in an edit error.)
XXX.9 Not documented in medical record
Ki-67 (MIB-1) not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is prior to 2021
Return to Schema ID Table

145 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00301: NET Duodenum (2018+)
See 00290: NET Stomach (2018+)
3863: Ki-67
00302: NET Ampulla of Vater (2018+)
See 00290: NET Stomach (2018+)
3863: Ki-67
00310: NET Jejunum and Ileum (2018+)
See 00290: NET Stomach (2018+)
3863: Ki-67
00320: NET Appendix (2018+)
See 00290: NET Stomach (2018+)
3863: Ki-67
00330: NET Colon and Rectum (2018+)
See 00290: NET Stomach (2018+)
3863: Ki-67
00340: NET Pancreas (2018+)
See 00290: NET Stomach (2018+)
3863: Ki-67
146 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
THORAX

147 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00360: Lung (2018+)
3929: Separate Tumor Nodules
Item Length: 1
NAACCR Item #: 3929
XML Parent-NAACCR ID: Tumor-separateTumorNodules
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00360: Lung (2018+)
Description
“Separate tumor nodules” refers to what is conceptually a single tumor with intrapulmonary metastasis
in the ipsilateral (same) lung. Their presence in the same or different lobes of lung from the primary
tumor affects the T and M categories.
Rationale
This data item was previously collected for Lung, SSF #1 and at least one standard setter is continuing to
collect it.
Definition
Separate tumor nodules are defined as intrapulmonary metastasis identified in the same lobe or same
lung (ipsilateral) originating from a single lung primary at the time of diagnosis. Biopsy of tumors may or
may not be performed. So long as there is a strong suspicion the multiple lesions are of the same
histological type by imaging, physician judgement, or microscopically, this meets the criteria of separate
tumor nodules representing intrapulmonary metastases. The presence of metastases to extrathoracic
sites does not change this distinction.
Coding guidelines
Record the presence of separate tumor nodules within the same ipsilateral lobe and/or different lobes
of the same lung which are considered a single primary. The histology of the separate tumors must be
the same. Histology may be determined clinically (presumed to be the same based on imaging or
physician judgement) or microscopically confirmed.
Code 0 when
o SINGLE TUMOR nodule only
o Separate tumor nodules present with DIFFERENT HISTOLOGIES
Code 1 when
o Separate tumor nodules present in the SAME LOBE with the SAME HISTOLOGY
Code 2 when
o Separate tumor nodules present in DIFFERENT LOBES of the SAME LUNG (ipsilateral)
with the SAME HISTOLOGY.
Code 3 when

148 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Separate tumor nodules present in SAME LOBE AND DIFFERENT LOBES of the SAME
LUNG with the SAME HISTOLOGY.
Code 4 when
o Separate tumor nodules present in SAME LUNG with the SAME HISTOLOGY and it’s
UNKNOWN IF they are in the SAME LOBE OR DIFFERENT LOBES.
Additional Information
Source documents: imaging reports and pathology reports
Coding Instructions and Codes
Note 1: Physician statement of Separate Tumor Nodules in the ipsilateral (same) lung can be used to
code this data item when no other information is available. See discussion of terminology in Note 4.
Separate tumor nodules in the contralateral lung are not coded in this data item.
Note 2: Code the presence and location of separate tumor nodules, also known as intrapulmonary
metastasis, at the time of diagnosis in this item. Separate tumor nodules can be defined clinically (by
imaging) and/or pathologically. They can be in the same or different lobes of the same lung as the
primary tumor. Their location is used to assign the T in the TNM system.
Note 3: For this item, only code separate tumor nodules of the same histologic type as the primary
tumor, also referred to as intrapulmonary metastases.
In the case of multiple tumor nodules determined to be the same primary, if not all nodules are
biopsied, assume they are the same histology
Note 4: Other situations that display multiple lesions are NOT coded in this item. Assign code 0 if the
multiple lesions belong to one of these other situations. Refer to the AJCC Staging System Lung for
standardized and precise definitions of the situations which aren’t separate tumor nodules. They are
second primary tumors, also called synchronous primary tumors (not the same histology as the
primary tumor)
multifocal lung adenocarcinoma with ground glass/lepidic features
diffuse pneumonic adenocarcinoma
Note 5: “Synchronous” describes the appearance in time compared to the primary tumor. Do not code
this item based solely on the word “synchronous”. If separate nodules are described as “metachronous,”
the nodules may be evidence of progression of disease in which case they would not be coded here.
Note 6: If there are multiple tumor nodules or foci and the terminology used is not readily identifiable as
one of the situations described in Note 4, consult with the pathologist or clinician. If no further
information is available, assign code 7 and DO NOT use the information to assign a T category or extent
of disease.
Note 7: Code 0 if relevant imaging or resection is performed and there is no mention of separate tumor
nodules.
Note 8: Code 9 if there is no relevant imaging or resection of the primary site.

149 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 No separate tumor nodules; single tumor only
Separate tumor nodules of same histologic type not identified/not present
Intrapulmonary metastasis not identified/not present
Multiple nodules described as multiple foci of adenocarcinoma in situ or minimally invasive
adenocarcinoma
Non-invasive neoplasm (behavior /2)
1 Separate tumor nodules of same histologic type in ipsilateral lung, same lobe
2 Separate tumor nodules of same histologic type in ipsilateral lung, different lobe
3 Separate tumor nodules of same histologic type in ipsilateral lung, same AND different lobes
4 Separate tumor nodules of same histologic type in ipsilateral lung, unknown if same or
different lobe(s)
7 Multiple nodules or foci of tumor present, not classifiable based on Notes 3 and 4
8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Separate Tumor Nodules not assessed or unknown if assessed
Return to Schema ID Table

150 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00360: Lung (2018+)
3937: Visceral and Parietal Pleural Invasion
Item Length: 1
NAACCR Item #: 3937
XML Parent-NAACCR ID: Tumor-visceralParietalPleuralInvasion
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00360: Lung (2018+)
Description
Visceral and Parietal Pleural Invasion is defined as invasion beyond the elastic layer or to the surface of
the visceral pleura.
Rationale
Visceral and Parietal Pleural Invasion (previously called “pleural/elastic layer invasion (PL)”) is a Registry
Data Collection Variable for AJCC. This data item was previously collected for Lung, SSF #2.
Definition
Invasion of one or more layers of the pleura covering the lung (visceral pleura), such as invasion beyond
the elastic layer of the pleura. The elastic layer may be identified on hematoxylin and eosin (H&E) stains
or by special stains looking for the elastic fibers. An elastic stain is not needed in most cases to assess
the pleura for invasion, only in those cases where the distinction between PL0 and PL1 is unclear on H&E
sections. Elastic stains may also be helpful in cases where the visceral and parietal pleura are adherent,
making it difficult to identify the boundary between the visceral pleural surface and the parietal pleura.
VPI is relevant for peripheral lung tumors. The presence of visceral pleural invasion by tumors smaller
than 3 cm changes the T category from pT1 to pT2 and increases the stage from IA to IB in patients with
no nodal disease or stage IIA to IIB in patients with peribronchial or hilar nodes. Studies have shown that
tumors smaller than 3 cm that penetrate beyond the elastic layer of the visceral pleura behave similarly
to similar-size tumors that extend to the visceral pleural surface. Visceral pleural invasion should
therefore be considered present not only in tumors that extend to the visceral pleural surface, but also
in tumors that penetrate beyond the elastic layer of the visceral pleura. Four to six layers of visceral
pleura may be described by the pathologist.
Coding guidelines
Record results of visceral pleural invasion as stated on pathology report. Do not code separate pleural
tumor foci or nodules in this field (discontinuous pleural metastasis).
Code 0 when
o No evidence of visceral and parietal pleural invasion or described as PL0
o Tumor does not penetrate beyond the elastic layer of the visceral pleura
o Extends to the elastic layer

151 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code 4 when
o Invasion of pleura without specifying visceral or parietal pleura
o Uncertain whether elastic stain has been performed to identify visceral pleura invasion
o Pathology report states PL1 or PL2
Code 5 when tumor extends to the parietal pleura (classified as T3) or described as PL3
Code 9 when
o No information in the medical record
o Only FNA performed
o Pathology report is not available
o Visceral and Parietal Pleural Invasion not evaluated (not assessed)
o Unknown if Visceral and Parietal Pleural Invasion evaluated (assessed)
Additional Information
Source documents: pathology report
For further information, refer to the Lung cancer protocol published by the College of American
Pathologists for the AJCC Staging System Lung
Other names: VPI, PL (number)
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of visceral pleural invasion, the registrar could assume that it was negative and
code appropriately. For the SSDI, this assumption cannot be made. There must be a statement
that visceral pleural invasion is not present to code 0
Coding Instructions and Codes
Note 1: Physician statement of Visceral and Parietal Pleural Invasion can be used to code this data item
when no other information is available.
Note 2: A surgical resection must be done to determine if the visceral and/or parietal pleural are
involved.
Note 3: Do not use imaging findings to code this data item
Note 4: Code 9 when
A FNA only is performed. A FNA is not adequate to assess pleural layer invasion
Surgical resection of the primary site is performed and there is no mention of visceral and/or
parietal pleural invasion
Code Description
0 No evidence of visceral pleural invasion identified
Tumor does not completely traverse the elastic layer of the pleura
Stated as PL0
Primary tumor is in situ
Non-invasive neoplasm (behavior /2)
No evidence of primary tumor
4 Invasion of visceral pleura present, NOS
Stated as PL1 or PL2
5 Tumor invades into or through the parietal pleura OR chest wall
Stated as PL3

152 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
6 Tumor extends to pleura, NOS; not stated if visceral or parietal
8
Not applicable:
Information not
collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
No surgical resection of primary site is performed
Visceral Pleural Invasion not assessed or unknown if assessed or cannot be determined
Return to Schema ID Table

153 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00360: Lung (2018+)
3938: ALK Rearrangement
Item Length: 1
NAACCR Item #: 3938
XML Parent-NAACCR ID: Tumor-alkRearrangement
Active years: 2021+
Schema(s):
00360: Lung (2018+)
Description
Testing for ALK rearrangement is performed for patients with advanced non-small cell lung cancer
(NSCLC) to identify tumors which are sensitive to small-molecule ALK kinase inhibitors.
Rationale
ALK rearrangement is recommended by treatment guidelines for patients with advanced lung cancer to
as a prognostic marker and factor in determining appropriate therapy. It is a new data item for cases
diagnosed 1/1/2021+.
Definition
“ALK positive cancer describes cancer cells that have a change in the structure of the anaplastic
lymphoma kinase (ALK) gene or a higher than normal amount of ALK protein on their surface. In normal
cells, ALK helps control cell growth. When cancer cells have the changed ALK gene or make too much
ALK protein, the cancer cells may grow more quickly. Knowing whether a cancer is ALK positive may help
plan treatment for advanced non-small cell cancers in the lung.” (NCI Dictionary of Cancer Terms
https://www.cancer.gov/publications/dictionaries/cancer-terms)
The presence of the ALK protein predicts a favorable response to therapy with a targeted ALK inhibitor,
such as crizotinib or ceritinib (chemotherapy).
Additional Information
Source documents: pathology report or clinical laboratory report, molecular report,
immunohistochemistry report
Other names: ALK tyrosine kinase receptor, anaplastic lymphoma kinase, anaplastic lymphoma
receptor tyrosine kinase, CD246, CD246 antigen, NBLST3
For further information, refer to the Lung Biomarker Reporting cancer protocol published by
the College of American Pathologists for the AJCC Staging System Lung
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of ALK rearrangement for non-small cell carcinoma can be used to code this
data item when no other information is available.
154 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
This data item only includes rearrangements. Ignore any amplifications or point mutations
Note 3: ALK may be recorded for all histologies and stages; however, it is primarily performed for
advanced non-small cell carcinomas. If information is not available, code 9.
Note 4: The absence or presence of ALK protein expression determines if the tumor will respond to
treatment with a targeted inhibitor. ALK protein expression predicts the ALK rearrangement gene, which
are more likely to respond to the targeted inhibitor treatment. The most common ALK rearrangements
are
EML4-ALK
KIF5B-ALK
TFG-ALK
KLC1-ALK
Note 5: If ALK Rearrangement is positive and there is no mention of the specific rearrangement, code 4.
Note 6: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no ALK results from pre-treatment specimens,
report the findings from post-treatment specimens
Note 7: Code 9 when
Insufficient amount of tissue available to perform test
Test done and documented to be equivocal
No microscopic confirmation of tumor
ALK Rearrangement not ordered or not done, or unknown if ordered or done
Code Description
0 Normal
ALK negative
Negative for rearrangement, no rearrangement identified, no mutations (somatic)
identified, not present, not detected
1 Abnormal Rearrangement identified/detected: EML4-ALK, KIF5B-ALK, TFG-ALK, and/or
KLC1-ALK
2 Rearrangement identified/detected: Other ALK Rearrangement not listed in code 1
4 Rearrangement, NOS
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9
Not documented in medical record
ALK Rearrangement not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is prior to 2021
Return to Schema ID Table

155 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00360: Lung (2018+)
3939: EGFR Mutational Analysis
Item Length: 1
NAACCR Item #: 3939
XML Parent-NAACCR ID: Tumor-egfrMutationalAnalysis
Active years: 2021+
Schema(s):
00360: Lung (2018+)
Description
Epidermal growth factor receptor (EGFR) mutational analysis is performed for patients with advanced
non-small cell lung cancer (NSCLC) to identify patients with certain activating mutations in the EGFR
gene which are sensitive to tyrosine kinase Inhibitors.
Rationale
EGFR mutational analysis is recommended by treatment guidelines for patients with advanced lung
cancer as a prognostic marker and factor in determining appropriate therapy. It is a new data item for
cases diagnosed 1/1/2021+.
Definition
“EGFR (epidermal growth factor receptor) is a protein found on certain types of cells that binds to a
substance called epidermal growth factor. The EGFR protein is involved in cell signaling pathways that
control cell division and survival. Sometimes, mutations (changes) in the EGFR gene cause EGFR proteins
to be made in higher than normal amounts on some types of cancer cells. This causes cancer cells to
divide more rapidly.” (NCI Dictionary of Cancer Terms
https://www.cancer.gov/publications/dictionaries/cancer-terms)
The presence of Exon 20 EGFR activating mutations are associated with a resistance to EGFR tyrosine
kinase inhibits, such as erlotinib, afatinib, and gefitinib. There is limited data available on response for
some of the other uncommon EGFR mutations (other than Exon 20). (CAP Cancer Protocol).
Additional Information
Source documents: pathology report or clinical laboratory report
Other names: Epidermal growth factor receptor tyrosine kinase inhibitor, ERBB, ERBB1, ErbB1,
HER1
For further information, refer to the Lung Biomarker Reporting cancer protocol published by
the College of American Pathologists for the AJCC Staging System Lung
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Physician statement of EGFR can be used to code this data item when no other information is
available.

156 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 3: EGFR may be recorded for all histologies and stages; however, it is primarily performed for
advanced non-small cell carcinomas. If information is not available, code 9.
Note 4: The most common EGFR mutations are
Exon 18 Gly719
Exon 19 deletion
Exon 20 insertion
Exon 20 Thr790Met
Exon 21 Leu858Arg
Note 5: If EGFR is positive and there is no mention of the mutated codon, or the mutated codon is not
specified, code 4.
Note 6: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no EGFR results from pre-treatment specimens,
report the findings from post-treatment specimens
Note 7: Code 9 when
Insufficient amount of tissue available to perform test
No microscopic confirmation of tumor
EGFR not ordered or not done, or unknown if ordered or done
Code Description
0 Normal
EGFR negative, EGFR wild type
Negative for mutations, no alterations, no mutations (somatic) identified, not present, not
detected
1 Abnormal (mutated)/detected in exon(s) 18, 19, 20, and/or 21
2 Abnormal (mutated)/detected but not in exon(s) 18, 19, 20, and/or 21
4 Abnormal (mutated)/detected, NOS, exon(s) not specified
7 Test ordered, results not in chart
8
Not
applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9
Not documented in medical record
EGFR not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is prior to 2021
Return to Schema ID Table

157 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00370: Pleura Mesothelioma (2018+)
3913: Pleural Effusion
Item Length: 1
NAACCR Item #: 3913
XML Parent-NAACCR ID: Tumor-pleuralEffusion
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00370: Pleural Mesothelioma (2018+)
Description
Pleural effusion is the accumulation of fluid between the parietal pleura (the pleura covering the chest
wall and diaphragm) and the visceral pleura (the pleura covering the lungs).
Rationale
Pleural Effusion can be collected by the surveillance community for pleura cancers. Prior to 2018, Pleura
SSF #1 was used for Pleural Effusion.
Definition
Pleural effusion is the accumulation of fluid between the two layers of pleura: visceral (covering the
lungs) and parietal (lining the chest wall and covering the diaphragm). Pleural effusion is a symptom of
mesothelioma that increases the Summary Stage from local or regional direct extension to distant
involvement.
Additional Information
Source documents: imaging, pathology and cytology reports
Other names: pleural fluid, thoracentesis
Coding guidelines
Record the absence or presence of pleural effusion. If pleural effusion is present and examined
microscopically, record whether the pleural effusion is non-malignant, malignant, or not specified.
Code 0 when there is no evidence of pleural effusion
Code 1 when
o Pleural effusion microscopically confirmed to be non-malignant
o Pleural effusion is stated to be negative for malignant cells
o Pleural effusion is seen on imaging, but pleural fluid cytology is negative for malignant
cells
Code 2 when
o Pleural effusion microscopically confirmed to be malignant
o Pleural effusion is stated to be positive for malignant cells
o Pleural fluid cytology described as suspicious or suspicious for mesothelioma
o Physician states pleural effusion is positive in the absence of positive cytology
Code 3 when

158 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Pleural fluid cytology is described as atypical or atypical mesothelial cells but not
specifically as non-malignant or malignant)
Code 4 when
o Pleural effusion is reported on imaging, but there is no cytology
o Pleural effusion is reported on imaging, but there is no physician’s statement on
whether it is positive
Code 9 when
o Not documented in the medical report
o Pleural effusion not evaluated (assessed)
o Unknown if Pleural Effusion evaluated (assessed)
Coding Instructions and Codes
Note 1: One of the most common symptoms of mesothelioma is a pleural effusion, or an accumulation
of fluid between the parietal pleura (the pleura covering the chest wall and diaphragm) and the visceral
pleura (the pleura covering the lungs). Record the absence or presence of pleural effusion and
specifically, if present, whether the pleural effusion is non-malignant, malignant, atypical or NOS.
Note 2: A physician’s statement of positive (malignant) pleural effusion or a positive cytology confirming
a malignant pleural effusion must be used to code this data item.
Code 2 when
o There is a positive malignant pleural effusion confirmed by cytology
o Pleural fluid cytology is described as suspicious/suspicious for mesothelioma
Code 3 when cytology is described as atypical/atypical mesothelial cells
Note 3: The presence of a pleural effusion on imaging alone (i.e., a pleural effusion, NOS) is not
equivalent to a malignant pleural effusion.
Code 1 if imaging indicates a pleural effusion but pleural cytology is described as negative for
malignant cells
Code 4 if imaging indicates a pleural effusion and there is no further information on whether it is
positive or negative or cytology not done
Code
Description
0 Pleural effusion not identified/not present
1 Pleural effusion present, non-malignant (negative)
2
Pleural effusion present, malignant (positive)
Physician states pleural effusion is malignant in the absence of positive cytology
3 Pleural effusion, atypical/atypical mesothelial cells
4 Pleural effusion, NOS
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Pleural effusion not assessed or unknown if assessed
Return to Schema ID Table
159 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
BONE

160 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00381: Bone Appendicular Skeleton (2018+)
3908: Percent Necrosis Post Neoadjuvant
Item Length: 5
NAACCR Item #: 3908
XML Parent-NAACCR ID: Tumor-percentNecrosisPostNeoadjuvant
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00381: Bone Appendicular Skeleton (2018+)
00382: Bone Spine (2018+)
00383: Bone Pelvis (2018+)
Description
Percent Necrosis Post Neoadjuvant is a prognostic factor for bone sarcomas.
Rationale
Percent Necrosis Post Neoadjuvant is a Registry Data Collection Variable for AJCC. It was previously
collected as Bone, CS SSF #3.
Definition
For osteosarcoma and Ewing’s sarcoma/PNET, response to neoadjuvant chemotherapy is a prognostic
factor. Patients with more than 90% tumor necrosis have a more favorable prognosis than those with
less response. The CAP protocol for bone tumor resection provides the pathologist with specific
instructions for determining the percentage of tumor necrosis. A separate method (system of Picci) may
describe response to treatment in grades: grade I (macroscopic viable tumor), grade II (microscopic
viable tumor), or grade III (no viable tumor). Do not code the Picci grade system in this data item.
Record the percentage value of tumor necrosis post neo-adjuvant chemotherapy as stated by the
pathologist in the pathology report. Code the value to the nearest whole percent in the range 001 to
100. If the patient has no resection or was not treated with pre-operative chemotherapy, code XXX.9
Additional Information
Source documents: pathology report
For further information, refer to the Bone cancer protocol published by the College of American
Pathologists for the AJCC Staging System Bone
Other names: Histologic treatment response, therapy response, chemotherapy effect
Coding instructions and Codes
Note 1: Physician statement of microscopically confirmed Percent Necrosis Post Neoadjuvant
Chemotherapy can be used to code this data item if no other documentation is available
Note 2: Record percentage value of the tumor necrosis post neoadjuvant chemotherapy as recorded in
the pathology report from resection of the primary tumor.

161 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 3: Code XXX.9 if
Surgical resection of the primary site after neoadjuvant therapy is performed and there is no
mention of percent necrosis
Surgical resection of the primary site is the initial therapy; therefore, no neoadjuvant therapy
was performed
Code Description
0.0 Tumor necrosis not identified/not present
0.1-
100.0
0.1 – 100.0 percent tumor necrosis
(Percentage of tumor necrosis to nearest tenth of a percent)
XXX.2 Tumor necrosis present, percent not stated
XXX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XXX.8 will result in an edit error.)
XXX.9 Not documented in medical record
No histologic examined of primary site
No neoadjuvant therapy
No surgical resection of primary site is performed
Return to Schema ID Table
163 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
SOFT TISSUE SARCOMA

164 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00400: Soft Tissue Head and Neck (2018+)
3815: Bone Invasion
Item Length: 1
NAACCR Item #: 3815
XML Parent-NAACCR ID: Tumor-boneInvasion
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00400: Soft Tissues Head and Neck (2018+)
00410: Soft Tissue Trunk and Extremities (2018+)
00421: Soft Tissues Abdomen and Thoracic (excluding Heart, Mediastinum, Pleura) (2018+)
00422: Heart, Mediastinum, and Pleura (2018+)
00440: Retroperitoneum (2018+)
00450: Soft Tissue Rare (2018+)
00459: Soft Tissue Other (2018+)
Description
Bone invasion, the presence or absence of bone invasion based on imaging, is a prognostic factor for
soft tissue sarcoma.
Rationale
Bone Invasion is a Registry Data Collection Variable in AJCC. This data item was previously collected for
Soft Tissue, SSF #3.
Definition
Direct tumor extension from the primary sarcoma into adjacent bone. This field does not include distant
or discontinuous metastases to the skeletal system. Information in this field is based on radiology and
other imaging techniques.
Coding guidelines
Code 0 when there is no evidence of bone invasion on imaging
Code 1 when there is evidence of bone invasion on imaging
Code 9 when
o No information in the medical record
o Bone invasion not evaluated (assessed)
o Unknown if bone invasion evaluated (assessed)
Additional Information
Source documents: imaging report
Coding Instructions and Codes
Note 1: Physician statement of Bone Invasion can be used to code this data item when no other
information is available.

165 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 2: Record bone invasion as determined by relevant imaging only for the primary tumor. Imaging
methodologies include computed tomography (CT) scans and magnetic resonance imaging (MRI).
Note 3: Code 0 if relevant imaging is performed and there is no mention of bone invasion.
Note 4: Code 9 if there is no relevant imaging of the primary site.
Code Description
0 Bone invasion not present/not identified on imaging
1 Bone invasion present/identified on imaging
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
Bone invasion not assessed or unknown if assessed
Return to Schema ID Table

167 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00410: Soft Tissue Trunk and Extremities (2018+)
3927: Schema Discriminator 2: Soft Tissue Sarcoma (C473, C475, C493-C495)
Item Length: 1
NAACCR Item #: 3927
XML Parent-NAACCR ID: Tumor-schemaDiscriminator2
NAACCR Alternate Name: None
Active years: 2018+
AJCC 8th Edition Chapter(s):
00410: Soft Tissue Trunk and Extremities (2018+)
00421: Soft Tissues Abdomen and Thoracic (excluding Heart, Mediastinum, Pleura) (2018+)
00459: Soft Tissue Other (2018+)
Definition
The ICD-O-3 assigned topography codes for the peripheral nerve and autonomic nervous systems
tumors (C47) and the connective, subcutaneous and other soft tissues (C49) primary sites are based on
transverse or horizontal planes. The AJCC Staging System Soft Tissue Sarcoma of the Trunk and
Extremities, and Soft Tissue Sarcoma of the Abdomen and Thoracic Visceral Organs base the eligible
sites as either external structures or internal viscera. For example
C493 axilla is an external site using the AJCC Staging System Soft Tissue Sarcoma of the Trunk
and Extremities
C493 axillary artery is an internal site using the AJCC Staging System Soft Tissue Sarcoma of the
Abdomen and Thoracic Visceral Organs
C475 sacrococcygeal region is a large area that may be either external or internal
o Need to determine the exact area involved to assign the correct chapter
o C475 external area of sacrococcygeal region uses the AJCC Staging System Soft Tissue
Sarcoma of the Trunk and Extremities
o C475 intrapelvic area of sacrococcygeal region uses the AJCC Staging System Soft Tissue
Sarcoma of the Abdomen and Thoracic Visceral Organs To develop a software algorithm
that can be used to send the registrar to the correct system/schema, this schema
discriminator was developed.
The schema discriminator is based on determining whether the structure involved is part of the external
structures or the internal viscera. This is accomplished by
Terms in ICD-O-3 topography codes sorted appropriately by the physician experts when possible
Instructions on what to do when terms are not specific enough to be assigned as external
structures or internal viscera
o Without additional information, these may not be staged, for example C475 pelvis
o With additional information, these may be determined to be external structures or
internal viscera
In addition to the topography codes and terms, there is also an option of “External sites, NOS”
and “Internal sites, NOS” for registrars to use to assign the schema discriminator. Registrars may
need to use additional information, including physician staging, to choose the appropriate
schema discriminator

168 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note 1: A schema discriminator is used to discriminate for peripheral nerve tumors (C473, C475) and
connective tissue tumors (C493, C494, C495) for the subsite in which the tumor arose.
Note 2: Code 1 is used for external structures and is assigned to the AJCC Staging System Soft Tissue
Sarcoma of the Trunk and Extremities (Schema ID 00410: Soft Tissue Sarcoma of the Trunk and
Extremities).
Example: Trapezius muscle (C493) is an external structure, on the outer layer or periphery of the
body
Note 3: Code 2 is used for internal structures and is assigned to the AJCC Staging System Soft Tissue
Sarcoma of the Abdomen and Thoracic Visceral Organs (Schema ID 00421: Soft Tissue Sarcoma of the
Abdomen and Thoracic Visceral Organs).
Example: Aorta (C493) is an internal structure, in the inner parts of the body
Note 4: Code 8 is only used for cases for 2018-2020 that have already been abstracted prior to the
Version 2.0 update (2021 update). It can also be used for 2018-2020 cases that are abstracted after the
2021 updates.
For cases diagnosed 2021+, code 8 cannot be used
Note 5: Code 9 is used for when there is not enough specific information to determine if the structure is
external or internal. These cases are collected in Schema ID 00459: Soft Tissue Other.
Example: Chest NOS (C493) does not provide enough information in order to determine if it is
either an external structure, on the outer layer or periphery of the body, or an internal
structure, in the inner parts of the body
Code Description Schema ID#/Description
1 External structures (sites), NOS
Examples of terms include:
Peripheral nerves and autonomic nervous system (C47)
Pelvis (C475)
o Buttock
o Gluteal region
o Groin
o Inguinal region
o Perineum
o Sacrococcygeal region (stated as
external)
Thorax (C473)
o Axilla
o Chest wall
o Infraclavicular region
o Scapular region
o Thoracic wall
Connective, subcutaneous and other soft tissues (C49)
00410: Soft Tissue Trunk and
Extremities

169 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description Schema ID#/Description
Abdomen (C494)
o Abdominal wall
o Abdominal wall muscle
o Iliopsoas muscle
o Psoas muscle
o Rectus abdominis muscle
o Umbilicus
Pelvis (C495)
o Buttock
o Gluteal region
o Gluteus maximus muscle
o Groin
o Inguinal region
o Perineum
o Sacrococcygeal region
Thorax (C493)
o Axilla
o Chest wall
o Infraclavicular region
o Intracostal muscle
o Latissimus dorsi muscle
o Pectoralis major muscle
o Scapular region
o Thoracic wall
o
Trapezius muscle
2 Internal structures and viscera (sites), NOS
Examples of terms include
Peripheral nerves and autonomic nervous system (C47)
Sacrococcygeal region (intrapelvic)
Connective, subcutaneous and other soft tissues (C49)
Abdomen (C494)
o Abdominal aorta
o Abdominal vena cava
o Celiac artery
o Inferior vena cava
o Mesenteric artery
o Renal artery
o Vena cava
Pelvis (C495)
o Iliac artery
o Iliac vein
Thorax (C493)
o Aorta
o Axillary artery
o Diaphragm
o Internal mammary artery
o
Subclavian artery
00421: Soft Tissue Abdomen
and Thoracic (excluding Heart,
Mediastinum, Pleura)

170 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description Schema ID#/Description
o Superior vena cava
o
Thoracic duct
8 Not applicable: Diagnosis date 2018-2020 00421: Soft Tissue Abdomen
and Thoracic (excluding Heart,
Mediastinum, Pleura)
9 Not specific enough to determine if external or internal
Examples of terms include
Peripheral nerves and autonomic nervous system (C47)
Pelvis (C475)
o Lumbosacral plexus
o Sacral nerve
o Sacral plexus
Thorax (C473)
o Chest
o Intercostal nerve
Connective, subcutaneous and other soft tissues (C49)
Thorax (C493)
o Chest, NOS
o
Thorax
00459: Soft Tissue Other
Return to Schema ID Table

171 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00421: Soft Tissue Abdomen and Thoracic (2018+)
See 00400: Soft Tissue Head and Neck (2018+)
3815: Bone Invasion
See 00410: Soft Tissue Trunk and Extremities (2018+)
3927: Schema Discriminator 2: Soft Tissue Sarcoma (C473, C475, C493-C495)
00422: Heart, Mediastinum and Pleura (2018+)
See 00400: Soft Tissue Head and Neck (2018+)
3815: Bone Invasion
Return to Schema ID Table

172 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00430: GIST (2018+)
3926: Schema Discriminator 1: Primary Peritoneum Tumor
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00430: GIST (2018+)
Definition
The GIST chapter includes a schema discriminator for C481 for location of the primary tumor because all
the peritoneum structures are coded to C481, but two separate stage tables are used to derive the TNM
values.
Coding Instructions and Codes
Note: Since both omental and peritoneal gastrointestinal stromal tumors (GIST) are coded with the
same ICD-O-3 topography code (C481), this data item must be used to identify the appropriate AJCC
stage table.
Code Description Stage Table
1
Mesentery
Mesoappendix
Mesocolon
Pelvic peritoneum
Rectouterine pouch
Cul de sac
Pouch of Douglas
Other specified peritoneal site
Small Intestinal, Esophageal,
Colorectal, Mesenteric and
Peritoneal GIST
2
Omentum
Gastric and Omental GIST
9
Unknown or no information
Not documented in medical record
Small Intestinal, Esophageal,
Colorectal, Mesenteric and
Peritoneal GIST
Return to Schema ID Table

173 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00430: GIST (2018+)
3865: KIT Gene Immunohistochemistry
Item Length: 1
NAACCR Item #: 3865
XML Parent-NAACCR ID: Tumor-kitGeneImmunohistochemistry
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00430: GIST (2018+)
Description
KIT Gene Immunohistochemistry (IHC) is the expression of the KIT gene in tumor tissue specimens based
on immunohistochemical (IHC) stains. A positive test is a diagnostic and predictive marker for GIST
tumors.
Rationale
KIT Gene Immunohistochemistry (IHC) is a Registry Data Collection Variable in AJCC. This data item was
previously collected for GIST schemas in CS (different SSFs).
Definition
KIT is a gene that regulates cell growth and differentiation. Mutations of this gene become oncogenes
and cause a gastrointestinal stromal tumor to ignore cellular control signals. About 85-90% of GIST
tumors contain oncogenic mutations of the KIT receptor gene. KIT immunohistochemistry is a special
immunofluorescent stain that turns mutated cells brown and confirms a diagnosis of GIST. The presence
of the KIT gene also indicates that the patient may respond to Gleevec or Sutent.
Additional Information
Source documents: pathology report (special stain)
Other names: CD117, c-kit receptor, KIT receptor tyrosine kinase, or SCFR (stem cell factor
receptor)
Coding Instructions and Codes
Note 1: Physician statement of KIT IHC can be used to code this data item when no other information is
available.
Note 2: KIT Gene Immunohistochemistry (IHC) is the expression of the KIT gene in tumor tissue
specimens based on immunohistochemical (IHC) stains. A positive test is a diagnostic and predictive
marker for GIST tumors. Do not record secondary or acquired mutations that may have developed
because of long-term imatinib treatment.
Note 3: Other names for KIT are CD117 or c-kit.
Note 4: Results from nodal or metastatic tissue may be used for KIT Gene Immunohistochemistry.

174 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 KIT negative/normal; within normal limits
1 KIT positive
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
Cannot be determined by pathologist
KIT not assessed or unknown if assessed
Return to Schema ID Table

175 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00440: Retroperitoneum (2018+)
See 00400: Soft Tissue Head and Neck (2018+)
3815: Bone Invasion
00458: Soft Tissue Rare (2018+)
See 00400: Soft Tissue Head and Neck (2018+)
3815: Bone Invasion
00459: Soft Tissue Other (2018+)
See 00400: Soft Tissue Head and Neck (2018+)
3815: Bone Invasion
See 00060: Cervical Lymph Nodes and Unknown Primary Tumors of the Head and Neck (2018+)
3926: Schema Discriminator 1: Occult Head and Neck Lymph Nodes
See 00410: Soft Tissue Trunk and Extremities (2018+)
3927: Schema Discriminator 2: Soft Tissue Sarcoma (C473, C475, C493-C495)
176 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
SKIN

177 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00460: Merkel Cell Carcinoma (2018+)
3830: Extranodal Extension Clin (non-Head and Neck)
Item Length: 1
NAACCR Item #: 3830
XML Parent-NAACCR ID: Tumor-extranodalExtensionClin
NAACCR Alternate Name: Extranodal Extension Clinical (non-Head and Neck)
Active years: 2018+
Schemas(s):
00460: Merkel Cell Skin (2018+)
00570: Penis (2018+)
Description
Extranodal Extension (ENE) Clinical is defined as “the extension of a nodal metastasis through the lymph
node capsule into adjacent tissue” during the diagnostic workup. This data item defines clinical ENE for
sites other than Head and Neck.
Rationale
Extranodal Extension Clinical (non-Head and Neck) is a Registry Data Collection Variable for AJCC. This
data item was previously collected for Penis, SSF #17.
Definition
The presence of extranodal extension (ENE) from regional lymph nodes is an important prognostic factor
in some cancers because these patients are rarely cured without some type of systemic chemotherapy
or radiation. Extranodal extension is defined as metastatic tumor growing from within the lymph node
outward through the lymph node capsule and into surrounding connective tissues.
This data item is for ENE that is detected clinically.
Coding guidelines
Code 0 when there are positive nodes clinically, but ENE not identified/not present.
Code 1 when there are positive nodes clinically, ENE is identified by physical exam WITH or
WITHOUT imaging
Code 2 when there are positive nodes clinically, ENE is identified by biopsy (microscopically
confirmed)
Code 7 when nodes are clinically negative (cN0)
Code 4 when there are positive nodes clinically, ENE is identified, but not known how identified
Code 9 when
o No information in the medical record
o Positive nodes clinically, not evaluated (assessed) for ENE
o Positive nodes clinically, unknown if evaluated (assessed) for ENE
o Lymph nodes not evaluated (assessed) clinically
o Unknown if lymph nodes evaluated (assessed) clinically

178 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Source documents: pathology report, imaging reports, physical exam
Other names: ENE, extracapsular extension, ECE
Coding Instructions and Codes
Note 1: Physician statement of Extranodal Extension (ENE) Clinical or physician clinical staging can be
used to code this data item when there is no other information available.
Note 2: Extranodal Extension Clinical is defined as “the extension of a nodal metastasis through the
lymph node capsule into adjacent tissue” identified during the diagnostic workup. ENE is the preferred
terminology. Other names include: extranodal spread, extracapsular extension, or extracapsular
spread.”
Note 3: Be aware that the rules for coding ENE for head and neck sites compared to non-head and neck
sites are different.
Note 4: Code the status of extranodal extension assessed during the diagnostic workup for the
assignment of the clinical stage for the most involved regional lymph node(s). This is mainly determined
by physical examination and includes statements such as fixed or matted nodes. Imaging may also be
used, as well as lymph node biopsies or sentinel node biopsies performed prior to any treatment. Do not
code ENE for any distant nodes.
Code Description
0 Regional lymph nodes involved, ENE not present/not identified during diagnostic workup
1 Regional lymph nodes involved, ENE present/identified during diagnostic workup, based on
physical exam WITH or WITHOUT imaging
2 Regional lymph nodes involved, ENE present/identified during diagnostic workup, based on
microscopic confirmation
4 Regional lymph nodes involved, ENE present/identified, unknown how identified
7
No lymph node involvement during diagnostic workup (cN0)
Non-invasive neoplasm (behavior /2)
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error)
9 Not documented in medical record
Clinical ENE not assessed or unknown if assessed during diagnostic workup
Clinical assessment of lymph nodes not done, or unknown if done
Return to Schema ID Table

179 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00460: Merkel Cell Carcinoma (2018+)
3833: Extranodal Extension Path (non-Head and Neck)
Item Length: 1
NAACCR Item #: 3833
XML Parent-NAACCR ID: Tumor-extranodalExtensionPath
NAACCR Alternate Name: Extranodal Extension Pathological (non-Head and Neck)
Active years: 2018+
Schemas(s):
00460: Merkel Cell Skin (2018+)
00570: Penis (2018+)
Description
Extranodal Extension (ENE) Pathological is defined as “the extension of a nodal metastasis through the
lymph node capsule into adjacent tissue. This data item defines pathological ENE for sites other than
Head and Neck.
Rationale
Extranodal Extension Pathological (non-Head and Neck) is a Registry Data Collection Variable for AJCC.
This data item was previously collected for Penis, SSF #17.
Definition
The presence of extranodal extension (ENE) from regional lymph nodes is an important prognostic factor
in some cancers because these patients are rarely cured without some type of systemic chemotherapy
or radiation. Extranodal extension is defined as metastatic tumor growing from within the lymph node
outward through the lymph node capsule and into surrounding connective tissues.
This data item is for ENE that is detected pathologically.
Coding guidelines
Code 0 when there are positive nodes pathologically, but ENE not identified/not present
Code 1 when there are positive nodes pathologically, ENE is identified
Code 7 when nodes are surgically resected, and they are negative (pN0)
Code 9 when
o No information in the medical record
o Positive nodes pathologically, not evaluated (assessed) for ENE
o Positive nodes pathologically, unknown if evaluated (assessed) for ENE
o Lymph nodes not evaluated (assessed) pathologically (no surgical resection of lymph
nodes)
o Unknown if lymph nodes evaluated pathologically (assessed)
Additional Information
Source documents: pathology report from surgical resection
Other names: ENE, extracapsular extension, ECE

180 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note 1: Physician statement of Extranodal Extension (ENE) Pathological or physician pathological staging
can be used to code this data item when there is no other information available.
Note 2: Extranodal extension is defined as "the extension of a nodal metastasis through the lymph node
capsule into adjacent tissue." ENE is the preferred terminology. Other names include: extranodal spread,
extracapsular extension, or extracapsular spread."
"A regional node extending into a distant structure or organ is categorized as ENE and is not
recorded as distant metastatic disease."
Note 3: Be aware that the rules for coding ENE for head and neck sites compared to non-head and neck
sites are different.
Note 4: Code the status of extranodal extension assessed on the surgical resection specimen for the
most involved regional lymph node(s). Do not code ENE for any distant lymph nodes. Code the status of
ENE based on the following criteria
Code 0
o Absence of ENE, positive lymph nodes assessed by lymph node dissection
o 1292: Scope of Regional Lymph Node Surgery must be 3-7
Code 1
o Presence of ENE assessed by Sentinel Lymph Node biopsy
o Presence of ENE assessed by lymph node dissection
o 1292: Scope of Regional Lymph Node Surgery must be 2-7
Code 7
o Lymph nodes negative for cancer assessed by Sentinel lymph node biopsy or lymph
node dissection
o 1292: Scope of Regional Lymph Node Surgery must be 2-7
Code 9
o Absence of ENE, positive lymph nodes assessed by Sentinel Lymph Node biopsy
A positive Sentinel Lymph Node biopsy cannot assess the absence of ENE, only
the presence of it. This is because there is not enough surrounding tissue in a
Sentinel Lymph node biopsy to accurately assess ENE
If codes 1 or 7 are used, this indicates that the lymph nodes were surgically resected or a
Sentinel Lymph Node biopsy was done and Scope of Regional Lymph Node Surgery [NAACCR
Data Item: 1292] must be 2-7
Code Description
0 Regional lymph nodes involved, ENE not present/not identified from surgical resection
1 Regional lymph nodes involved, ENE present/identified from surgical resection
7 No lymph node involvement from surgical resection (pN0)
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error)
9 Not documented in medical record
No surgical resection of regional lymph nodes
Non-invasive neoplasm (behavior /2)
Cannot be determined
Pathological assessment of lymph nodes not done, or unknown if done
Extranodal Extension Pathological not assessed or unknown if assessed
Return to Schema ID Table

181 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00460: Merkel Cell Carcinoma (2018+)
3880: LN Isolated Tumor Cells (ITC)
NAACCR Item #: 3880
XML Parent-NAACCR ID: Tumor-lnIsolatedTumorCells
NAACCR Alternate Name: Lymph Nodes Isolated Tumor Cells (ITC)
Active years: 2018+
Schemas(s):
00460: Merkel Cell Skin (2018+)
Description
Lymph Nodes Isolated Tumor Cells (ITC), the presence of isolated tumor cells in regional lymph node(s)
that may be detected by hematoxylin and eosin or by immunohistochemical staining, is a potential
prognostic factor for Merkel Cell Carcinoma.
Rationale
Lymph Nodes, Isolated Tumor Cells (ITC) is a Registry Data Collection Variable in AJCC. This data item
was previously collected for Merkel Cell Skin, SSF #18.
Definition
Isolated tumor cells (ITCs) for Merkel cell carcinoma are defined as single tumor cells or small clusters of
tumor cells not more than 0.2 mm in greatest dimension. ITCs are usually detected by
immunohistochemistry on sentinel lymph node biopsies.
Note: Examples of immunohistochemical staining methods are Cytokeratin 20 (CK20), CAM 5.2,
pancytokeratin, and AE1/3. ITCs may be detected by routine H&E stains.
Additional information
Source documents: pathology report
Coding Instructions and Codes
Note 1: Physician statement of Isolated Tumor Cells (ITCs) can be used to code this data item when no
other information is available.
Note 2: ITCs include single tumor cells or small clusters, less than or equal to 0.2 mm in greatest
dimension, generally without stromal response in the lymph node. These cells usually are found in the
subcapsular nodal sinuses but may be seen within the nodal parenchyma.
Note 3: ITCs may be identified in lymph nodes by hematoxylin and eosin staining or by specialized
pathological techniques, such as IHC for cytokeratin proteins for carcinomas. Specialized pathology
techniques such IHC and molecular techniques are not recommended for routine examination of lymph
nodes.
Note 4: Record the status of ITCs as documented by the pathologist.

182 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Regional lymph nodes negative for ITCs
1
Regional lymph nodes positive for ITCs
(Tumor cell clusters not greater than 0.2 millimeter (mm))
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
Cannot be determined by pathologist
ITCs not assessed or unknown if assessed
Return to Schema ID Table

183 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00460: Merkel Cell Carcinoma (2018+)
3918: Profound Immune Suppression
Item Length: 1
NAACCR Item #: 3918
XML Parent-NAACCR ID: Tumor-profoundImmuneSuppression
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00460: Merkel Cell Skin (2018+)
Description
Profound Immune Suppression, suppressed immune status that may be associated with HIV/AIDs, solid
organ transplant, chronic lymphocytic leukemia, non-Hodgkin lymphoma, multiple conditions or other
conditions, increases the risk of developing Merkel Cell Carcinoma and is an adverse prognostic factor.
Rationale
Profound Immune Suppression is a Registry Data Collection Variable in AJCC. It was previously collected
as Merkel Cell Penis, SSF #22, Merkel Cell Scrotum SSF #22, Merkel Cell Skin, SSF #22, and Merkel Cell
Vulva, SSF #22.
Definition
Profound immune suppression may greatly increase the risk of developing Merkel cell carcinoma.
Immune suppression is suppression of the body's immune system and its ability to fight infections and
other diseases. Immune suppression may be deliberately induced with drugs, as in preparation for bone
marrow or other organ transplantation, to prevent rejection of the donor tissue. It may also result from
certain diseases such as Acquired Immune Deficiency Syndrome (AIDS) or lymphoma, and from the use
of anti-cancer drugs.
Additional Information
Source documents: patient history, consultation notes, other statement in medical record
Other names: immunosuppression
Coding Instructions and Codes
Note 1: Physician statement of Profound Immune Suppression must be used to code this data item. Do
not assume that a patient is immune suppressed just because the patient has one of the conditions
listed below in the table. Per AJCC experts, the following terms can also be used to describe “profound
immune suppression.”
Immunocompromised
Immunosuppressed
Suppressed immune status
Note 2: Per AJCC experts, this data item is limited to the conditions in the table below occurring within
two years of the diagnosis of Merkel cell carcinoma.

184 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
For the following conditions, these patients will experience chonic immunosuppression. These
are no time limits for these conditions. If a patient has a history (regardless of when diagnosed
or treatment status), code as present
o Human Immunodeficiency Virus (HIV)/Acquired Immunodeficiency Syndrome (AIDS)
(Code 1)
o Solid organ transplant recipient (Code 2)
o Chronic lymphocytic leukemia (Code 3)
Note 3: Code 9 if conditions in the table below were not active within 2 years of (or resolved more than
2 years prior to) diagnosis, or if it is unknown when they existed.
Note 4: If more than one condition is documented, code 5. Document the specific conditions in the text
field.
Code Description
0 No immune suppression condition(s) identified/not present
1 Human Immunodeficiency Virus (HIV)/Acquired Immunodeficiency Syndrome (AIDS)
2 Solid organ transplant recipient
3 Chronic lymphocytic leukemia
4 Non-Hodgkin lymphoma
5 Multiple immune suppression conditions
6
Profound immune suppression present
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9
Not documented in medical record
Profound immune suppression not assessed or unknown if assessed
Return to Schema ID Table

185 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3817: Breslow Tumor Thickness
Item Length: 4
NAACCR Item #: 3817
XML Parent-NAACCR ID: Tumor-breslowTumorThickness
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00470: Melanoma Skin (2018+)
Description
Breslow Tumor Thickness, the measurement of the thickness of a melanoma as defined by Dr. Alexander
Breslow, is a prognostic factor for Melanoma of the Skin
Rationale
Breslow Tumor Thickness is a Registry Data Collection Variable in AJCC. It was previously collected as
Melanoma Skin, CS SSF #1.
Definition
A measure of how deeply a melanoma tumor has grown into the skin. The tumor thickness (depth) is
usually measured from the top of the tumor to the deepest tumor cells. If the tumor is ulcerated (the
skin is broken), it is measured from the base of the ulcer to the deepest tumor cells. Breslow thickness is
used to help determine the stage of cancer. Thicker tumors are linked with lower survival rates.
Coding guidelines
Code a measurement specifically labeled as “thickness” or “depth” or “Breslow depth of invasion” from
the pathology report. In the absence of this label, a measurement described as taken from the cut
surface of the specimen may be coded. And in the absence of either of these labels, the third dimension
in a statement of tumor size can be used to code this field.
Code the greatest measured thickness from any procedure performed on the lesion, whether it is
described as a biopsy or an excision. Do not add measurements together from different procedures.
Example: A punch biopsy with a thickness of 0.5 mm is followed by a re-excision with a thickness
of residual tumor of 0.2 mm. Code 0.5 mm.
If the tumor is excised post-neoadjuvant treatment, tumor measurements cannot be compared before
and after treatment to determine which would indicate the greater involvement. The same code (XX.9)
is used for cases with no surgical procedure of the primary site and cases with surgical procedure of the
primary site after neoadjuvant treatment.
Because the thickness table is similar to many other tables that collect a measurement, it is important to
identify the correct unit of measurement.

186 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
In the range 0.1-99.9, code the actual tumor thickness, tumor depth, or Breslow measurement in tenths
of millimeters as stated in the pathology report. If the measurement is given in hundredths of
millimeters, use the general rules for rounding to determine the value in tenths of millimeters. This is a
four-digit field with a decimal point in the third digit.
Examples: Tumor described as 0.5 mm in depth – code as 0.5. Lesion 1 mm thick – code as
1.0. Breslow 2.5 mm – code as 2.5. Thickness of 10 mm (1 cm) – code as 10.0).
Additional Information
Source documents: pathology report
For further information, refer to the Skin Melanoma cancer protocol published by the College of
American Pathologists for the AJCC Staging System Melanoma of the Skin
Other names: maximum tumor thickness, Breslow depth of invasion, Breslow thickness, Breslow
measurement, Breslow’s microstaging
Coding Instructions and Codes
Note 1: Physician statement of Breslow Tumor Thickness can be used to code this data item when no
other information is available, or the available information is ambiguous.
Note 2: Code Breslow tumor thickness, not size. Record actual measurement in tenths of millimeters
from the pathology report. Measurement given in hundredths of millimeters should be rounded to the
nearest tenth.
Examples:
0.4 mm – 0.4
1.0 mm- 1.0
2.5 mm – 2.5
2.56 mm- 2.6
11 mm – 11.0
12.35 mm – 12.4 mm
Note 3: Code the greatest measured thickness from any procedure performed on the lesion, whether it
is described as a biopsy or an excision.
For example, if a punch biopsy with a thickness of 1.5 mm is followed by a re-excision with a
thickness of residual tumor of 0.2 mm, code 1.5.
Note 4: If there are multiple procedures and the pathologist adds the measurement together to get a
final Breslow’s depth, the registrar can use this.
Do not add the measurements together, only the pathologist can do this
Note 5: If the pathologist describes the thickness as “at least,” use the appropriate A code. An exact
measurement takes precedence over A codes.
If the pathologist states “greater than” instead of “at least”, code to XX.9, unless it is greater
than 9.9 mm (Code AX.0)
Examples:
Pathologist states the thickness is “at least 2.0 mm.” Code A2.0
Pathologist states the thickness is “greater than 4 mm.” Code XX.9

187 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0.0 No mass/tumor found
0.1 Greater than 0.0 and less than or equal to 0.1
0.2-99.9 0.2 – 99.9 millimeters
XX.1 100 millimeters or larger
A0.1
-
A9.9
Stated as “at least” some measured value of 0.1 to 9.9
AX.0 Stated as greater than 9.9 mm
XX.8
Not applicable: Information not collected for this schema
(If this item is required by your standard setter, use of code XX.8 will result in an edit error)
XX.9 Not documented in medical record
Microinvasion; microscopic focus or foci only and no depth given
Cannot be determined by pathologist
Non-invasive neoplasm (behavior /2)
Breslow Tumor Thickness not assessed or unknown if assessed
Return to Schema ID Table

188 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3936: Ulceration
Item Length: 1
NAACCR Item #: 3936
XML Parent-NAACCR ID: Tumor-ulceration
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00470: Melanoma Skin (2018+)
Description
Ulceration, the absence of an intact epidermis overlying the primary melanoma based upon
histopathological examination, is a prognostic factor for melanoma of the skin.
Rationale
Ulceration is a Registry Data Collection Variable in AJCC. It was previously collected as Melanoma Skin,
CS SSF #2.
Definition
Ulceration is the formation of a break on the skin or on the surface of an organ. An ulcer forms when the
surface cells die and are cast off. Ulcers may be associated with cancer and other diseases.
Primary tumor ulceration has been shown to be a dominant independent prognostic factor, and if
present, changes the pT stage from T1a to T1b, T2a to T2b, etc., depending on the thickness of the
tumor.
The presence or absence of ulceration must be confirmed on microscopic examination. Melanoma
ulceration is defined as the combination of the following features
Full-thickness epidermal defect (including absence of stratum corneum and basement
membrane)
Evidence of reactive changes (i.e., fibrin deposition, neutrophils); and thinning, effacement, or
reactive hyperplasia of the surrounding epidermis in the absence of trauma or a recent surgical
procedure
Ulcerated melanomas typically show invasion through the epidermis, whereas nonulcerated
melanomas tend to lift the overlying epidermis
Coding guidelines
Record whether ulceration is present or absent
Code 0 when there is a statement in the pathology report that no ulceration is present
Code 1 when the pathologist states that ulceration is present
Code 9 when
o No information in the medical record
o Pathology report is not available
o Ulceration not evaluated (not assessed)
o Unknown if Ulceration evaluated (assessed)

189 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Source documents pathology report, physical exam, consultant notes, other statement in
medical record
For further information, refer to the Skin Melanoma cancer protocol published by the College of
American Pathologists for the AJCC Staging System Melanoma of the Skin
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of ulceration, the registrar could assume that it was negative and code
appropriately. For the SSDI, this assumption cannot be made. There must be a statement that
ulceration is not present to code 0
Coding Instructions and Codes
Note 1: Physician statement of microscopically confirmed ulceration (e.g., based on biopsy or surgical
resection) can be used to code this data item.
Note 2: Ulceration can only be confirmed by microscopic examination. Do not use findings from physical
exam.
It is possible for a patient to present with an ulcerated lesion noted on physical exam, but this is
not the same thing as ulceration seen on a microscopic exam
Note 3: Melanoma ulceration is the absence of an intact epidermis overlying the primary melanoma
based upon histopathological examination.
Code 1 if any biopsy (punch, shave, excisional, etc.) or wide excision is positive for ulceration in
the presence of an underlying melanoma
Code 0 is all specimens are negative OR one specimen is negative and the other is unknown
Ulceration must be caused by an underlying melanoma. Ulceration caused by trauma from a
previous procedure should not be coded as positive for this SSDI
Note 4: Code 9 if there is microscopic examination and there is no mention of ulceration.
This instruction does apply to non-invasive neoplasms (behavior 2)
Code
Description
0 Ulceration not identified/not present
1 Ulceration present
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Cannot be determined by the pathologist
Pathology report does not mention ulceration
Ulceration not assessed or unknown if assessed
Return to Schema ID Table

190 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3893: Mitotic Rate Melanoma
Item Length: 2
NAACCR Item #: 3893
XML Parent-NAACCR ID: Tumor-mitoticRateMelanoma
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00470: Melanoma Skin (2018+)
Description
Mitotic Rate Melanoma, the number of mitoses per square millimeter based on pathological evaluation,
is a prognostic factor for melanoma of the skin.
Rationale
Mitotic Rate Melanoma is a Registry Data Collection Variable in AJCC. It was previously collected as
Melanoma Skin, CS SSF #7.
Definition
Mitotic count is a way of describing the potential aggressiveness of a tumor. Record the number of cells
actively dividing as determined by the pathologist. The count will vary according to the type of tumor.
Additional Information
Source documents: pathology report
For further information, refer to the Skin Melanoma cancer protocol published by the College of
American Pathologists for the AJCC Staging System Melanoma of the Skin
Other names: mitotic rate, mitotic index (a ratio—do not record this measurement), mitotic
activity
Coding Instructions and Codes
Note 1: Physician statement of the Mitotic Rate Melanoma can be used to code this data item when no
other information is available.
Note 2: The term “mitotic figures” is the same as mitoses.
Note 3: Record the mitotic rate/count as documented in the pathology report. If there is more than one
pathology report for the same melanoma at initial diagnosis and different mitotic counts are
documented, code the highest mitotic count from any of the pathology reports.

191 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
00 0 mitoses per square millimeter (mm)
Mitoses absent
No mitoses present
01-99 1 - 99 mitoses/square mm
(Exact measurement in mitoses/square mm)
X1 100 mitoses/square mm or more
X2 Stated as "less than 1 mitosis/square mm"
Stated as "nonmitogenic"
X3 Stated as "at least 1 mitosis/square mm"
Stated as "mitogenic"
X4 Mitotic rate described with denominator other than square millimeter (mm)
X7 Test ordered, results not in chart
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit error.)
X9 Not documented in medical record
Mitotic Rate Melanoma not assessed or unknown if assessed
Return to Schema ID Table

192 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3932: LDH Lab Value
Item Length: 7
NAACCR Item #: 3932
XML Parent-NAACCR ID: Tumor-ldhPretreatmentLabValue
NAACCR Alternate Name: LDH (Lactate Dehydrogenase) Lab Value, LDH (Lactate Dehydrogenase) Lab
Value
Active years: 2018+
Schemas(s):
00470: Melanoma Skin (2018+)
Description
LDH (Lactate Dehydrogenase) Lab Value, measured in serum, is a predictor of treatment response,
progression-free survival and overall survival for patients with Stage IV melanoma of the skin.
Rationale
LDH Lab Value is a Registry Data Collection Variable in AJCC. It was previously collected as Melanoma
Skin, CS SSF #5.
Definition
When cells (normal or tumor) are damaged or destroyed, an enzyme called lactate dehydrogenase (LDH)
is released into the bloodstream. LDH is an indirect indication of possible tumor burden or damage to
an organ, which may be caused by metastatic involvement of liver or lung, or a myocardial
infarction. The total LDH should be the test value that is coded, but there are five fractions of LDH that
measure tissue specific cellular damage: LD1 and LD2: heart, red blood cells and kidneys; LD3: lung; LD4
and LD5: liver, skin and skeletal muscles. LDH is elevated in 60% of patients with non-seminomatous
germ cell tumors of the testis. LDH is not a screening test, nor is it diagnostic of melanoma, ocular
adnexal lymphoma, or testicular cancer.
Coding guidelines
Code 0.0 for a test result of 0 (U/L).
Code the highest exact LDH lab value prior to systemic (chemo, immunotherapy, hormone),
radiation therapy or surgery to a metastatic site in the range 0.1 to 99,999.9
Code XXXXX.1 for a total LDH lab value of 100,000 or greater.
Code XXXXX.7 if the test was ordered and the results are not in the medical record.
Code XXXXX.9 when
o there is no information in the medical record about the LDH lab value
o Test is not done or unknown if the test was done
Additional Information
Source documents: clinical laboratory report; may be included in a liver or hepatic panel/profile,
a cardiac panel, or a general metabolic panel of tests

193 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Other names: LDH, Lactate dehydrogenase, lactase dehydrogenase, lactic acid dehydrogenase
Coding Instructions and Codes
Note 1: Physician statement of LDH (Lactate Dehydrogenase) Lab Value can be used to code this data
item when no other information is available.
Note 2: LDH is important in melanoma staging in the setting of DISTANT metastasis. LDH level might
only be ordered after re-excision/wide excision and/or nodal evaluation indicates a higher risk of distant
metastasis. Imaging may then be performed and if distant metastasis are identified, LDH is ordered.
Note 3: Record the lab value of the highest serum LDH test results documented in the medical record
either before or after surgical resection of the primary tumor with or without regional lymph node
dissection. The LDH must be taken prior to systemic (chemo, immunotherapy, hormone), radiation
therapy or surgery to a metastatic site. The lab value may be recorded in a lab report, history and
physical, or clinical statement in the pathology report.
Note 4: The same laboratory test should be used to record information in 3869: LDH Level and 3870:
LDH Upper Limits of Normal.
Code
Description
0.0 0.0 (U/L)
0.1
-
99999.9
0.1
–
99,999.9 U/L
XXXXX.1 100,000 U/L or greater
XXXXX.7 Test ordered, results not in chart
XXXXX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XXXXX.8 will result in an edit error.)
XXXXX.9 Not documented in medical record
LDH (Lactate Dehydrogenase) Lab Value not assessed or unknown if assessed
Return to Schema ID Table

194 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3869: LDH Level
Item Length: 1
NAACCR Item #: 3869
XML Parent-NAACCR ID: Tumor-ldhPretreatmentLevel
NAACCR Alternate Name: LDH (Lactate Dehydrogenase) Level, LDH (Lactate Dehydrogenase) Level
Active years: 2018+
Schemas(s):
00470: Melanoma Skin (2018+)
00821: Plasma Cell Myeloma (2018+)
Description
LDH (Lactate Dehydrogenase) is an enzyme involved in conversion of sugars to energy and present in
most cells in the body. Elevated LDH is an adverse prognostic factor for plasma cell myeloma and
melanoma of the skin.
Rationale
LDH (Lactate Dehydrogenase) Level is a prognostic factor required in AJCC 8
th
edition for Chapter 82
Plasma Cell Myeloma and Plasma Cell Disorders and Chapter 47 Melanoma Skin. For Plasma Cell
Myeloma, LDH is part of the RISS Stage and is new for cases diagnosed 1/1/2018+. For Melanoma Skin,
LDH is used to define the M subcategories and was previously collected as Melanoma Skin, SSF #4.
Coding Instructions and Codes
Note 1: Use the reference ranges from your lab to determine if LDH is normal.
Note 2: Record the lab value of the highest serum LDH test results documented in the medical record
either before or after surgical resection of the primary tumor with or without regional lymph node
dissection. The LDH must be taken prior to systemic (chemo, immunotherapy, hormone), radiation
therapy or surgery to a metastatic site. The lab value may be recorded in a lab report, history and
physical, or clinical statement in the pathology report.
Note 3: If there is no mention of the LDH, code 9.
Note 4: The same laboratory test should be used to record information in 3870: LDH Upper Limits of
Normal and 3932: LDH Lab Value.
Code Description
0
Normal LDH level
Low, below normal
1 Above normal LDH level; High
7 Test ordered, results not in chart
9 Not documented in medical record
LDH (Lactate Dehydrogenase) Level not assessed or unknown if assessed
Return to Schema ID Table

195 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3870: LDH Upper Limits of Normal
NAACCR Item #: 3870
XML Parent-NAACCR ID: Tumor-ldhUpperLimitsOfNormal
NAACCR Alternate Name: LDH (Lactate Dehydrogenase) Upper Limits of Normal
Active years: 2018+
Schemas(s):
00470: Melanoma Skin (2018+)
Description
LDH (Lactate Dehydrogenase), an enzyme involved in converting sugars to energy in the body, is
elevated in some malignancies. LDH level is a prognostic factor for patients with Stage IV melanoma.
This data Item refers to the Upper Limit of Normal in the laboratory test used to interpret the Serum
LDH result.
Rationale
LDH (Lactate Dehydrogenase) Upper Limits of Normal is a Registry Data Collection Variable in AJCC. It
was previously collected as Melanoma Skin, CS SSF #6.
Definition
When cells (normal or tumor) are damaged or destroyed, an enzyme called lactate dehydrogenase (LDH)
is released into the bloodstream. LDH is an indirect indication of possible tumor burden or damage to
an organ, which may be caused by metastatic involvement of liver or lung, or a myocardial
infarction. The total LDH should be the test value that is coded, but there are five fractions of LDH that
measure tissue specific cellular damage: LD1 and LD2: heart, red blood cells and kidneys; LD3: lung; LD4
and LD5: liver, skin and skeletal muscles. LDH is elevated in 60% of patients with non-seminomatous
germ cell tumors of the testis. LDH is not a screening test, nor is it diagnostic of melanoma, ocular
adnexal lymphoma, or testicular cancer.
Additional Information
Source documents: clinical laboratory report; may be included in a liver or hepatic panel/profile,
a cardiac panel, or a general metabolic panel of tests
Other names: LDH, Lactate dehydrogenase, lactase dehydrogenase, lactic acid dehydrogenase
Normal reference range: varies widely by laboratory, patient age, and the units of
measurement.
Examples of reference range lab values:
o Lab A Total LDH 71 – 207 U/L
o Lab B Total LDH 300 – 600 U/L
o Lab C Total LDH 45 – 90 U/L
o Lab D Total LDH 150 – 250 U/L
Coding Instructions and Codes

196 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 1: Physician statement of LDH (Lactate Dehydrogenase) Upper Limit of Normal can be used to code
this data item.
Note 2: Record the value of the highest serum LDH test results documented in the medical record either
before or after surgical resection of the primary tumor with or without regional lymph node dissection.
The LDH must be taken prior to systemic (chemo, immunotherapy, hormone), radiation therapy or
surgery to a metastatic site. The lab value may be recorded in a lab report, history and physical, or
clinical statement in the pathology report.
Note 3: Upper limits of normal for LDH vary widely depending on the lab. Common upper limits can be
200, 250, 618, or other values.
Note 4: The same laboratory test should be used to record information in 3932: LDH Lab Value and
3869: LDH Level.
Code Description
001-
999
001 - 999 upper limit of normal
(Exact upper limit of normal)
XX8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XX8 may result in an edit error.)
XX9 Not documented in medical record
LDH Upper Limit not assessed or unknown if assessed
Return to Schema ID Table

197 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00470: Melanoma Skin (2018+)
3961: Clinical Margin Width
Item Length: 4
NAACCR Item #: 3961
XML Parent-NAACCR ID: clinicalMarginWidth
NAACCR Alternate Name: None
Active years: 2023+
Schemas(s):
00470: Melanoma Skin (2018+)
Description
Clinical margin width describes the margins from a wide excision for a melanoma primary. The margin
width is measured by the surgeon prior to the procedure. The measurement is taken, in centimeters,
from the edge of the lesion or the prior excision scar to the peripheral margin of the specimen.
Rationale
Clinical margin width for wide local excision of a melanoma is based on the original Breslow thickness of
the primary tumor, as indicated on the initial biopsy pathology report.
Definition
Per the American College of Surgeons Optimal Resources for Cancer Care-2020 Standards Standard 5.5
Local Excision for Primary Cutaneous Melanoma, the clinical margin width for wide local excision of
invasive melanoma should be 1 cm for melanomas <1 mm thick, 1 to 2 cm for invasive melanomas 1 to 2
mm thick, and 2 cm for invasive melanomas >2 mm thick. The clinical margin width for wide local
excision of a melanoma in situ should be at least 5 mm.
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2023+
For cases diagnosed 2018-2022, leave this SSDI blank
Note 2: “The appropriate [wide local excision] margins are measured from the periphery of any gross
residual tumor or the edges of the entire previous biopsy scar (shave or excisional).” Operative
Standards for Cancer Surgery, Volume 2, page 392.
Note 3: Code the peripheral surgical margins from the operative report from a wide excision
Do not use the pathology report to code this data item.
Margins from wide excision-Measured from the edge of the lesion or the prior excision scar to
the peripheral margin of the specimen, do not use deep margin
Do not add margins together
If multiple wide excisions are performed, code the clinical margin width from the procedure
with the largest margin
Note 4: Physician statement of clinical margin width can be used to code this data item when no other
information is available, or the available information is ambiguous
Order of priority

198 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Operative Note
Physician statement in medical record
Note 5: Record stated margin in centimeters. Include decimal point.
Examples:
0.5 cm - 0.5
1 cm- 1.0
.5 cm - 2.5
Code
Description
0.1 Documented as 0.1cm or less (1mm or less)
0.2-9.9 0.2 cm – 9.9 cm
XX.1 10 centimeters or greater
XX.8 Not Applicable. Information not collected for this schema
(If this information is required by your standard setter, use of code XX.8 may
result in an edit error)
XX.9 Not documented in medical record
No Wide Excision performed
Mohs or similar procedure
Wide Excision performed, but clinical margin width not documented.
No surgical resection performed (B000)
Unknown if procedure performed.
BLANK N/A-Diagnosis year is prior to 2023.
Return to Schema ID Table
199 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
BREAST

200 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
Estrogen Receptor and Progesterone Receptor
Definition
Estrogen receptor (ER) positivity and progesterone receptor (PR) positivity are favorable prognostic
factors in breast cancer, as well as endometrial carcinoma and meningioma. Positive results predict a
favorable response to endocrine (hormonal) therapy. Combined ER and PR positivity is associated with
increased response to antiestrogen therapies.
There are a variety of ways to report information on ER and PR results, but there is almost always a
summary statement that the result is positive or negative.
The following data items are used to collect ER and PR information
3826: Estrogen Receptor Percent Positive or Range
3827: Estrogen Receptor Summary
3828: Estrogen Receptor Total Allred Score
3914: Progesterone Receptor Percent Positive or Range
3915: Progesterone Receptor Summary
3916: Progesterone Receptor Total Allred Score
Note: Do not use results from the following tests to record ER or PR results
Oncotype Dx
MammaPrint
EndoPredict
PAM 50 (Prosigna)
Any other test that records HER2
The two most common ways to report ER and PR results are the percentage of cells with nuclear
positivity and the average intensity of staining. Both the PS and IS are based on
immunohistochemical staining of tumor cells.
ER and PR status, the percentage of tumor cells with positive nuclear staining, may be reported as a
specific number or a range if more than 10%. Intensity refers to degree of nuclear positivity (i.e.,
pale to dark); average intensity of staining is recorded as weak, moderate or strong.
ER or PR Status
___ Positive
Percentage of cells with nuclear positivity
#
Specify: ___ %
-OR-
Range (Note A)
___ 1-10% (specify): ____ %
#
___ 11-20%
___ 21-30%
___ 31-40%
___ 41-50%
___ 51-60%

201 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
___ 61-70%
___ 71-80%
___ 81-90%
___ 91-100%
+ Average intensity of staining:
+ ___ Weak
+ ___ Moderate
+ ___ Strong
___ Negative
Allred Score for Estrogen and Progesterone Receptor Evaluation
The Allred Score is a method of quantifying ER and PR using both intensity and percentage of positive
cells. The Allred Score is calculated by adding the Proportion Score and the Intensity Score, as defined in
the tables below.
The Allred score combines the percentage of positive cells (proportion score) and the intensity score of
the reaction product in most of the carcinoma. The 2 scores are added together for a final score with 8
possible values (00-08).
Proportion Score Positive Cells, %
0 0
1 <1
2 1 to 10
3
11 to 33
4
34 to 66
5
Intensity Intensity Score
None 0
Weak 1
Intermediate/Moderate
2
Strong 3
Additional Information
For further information, refer to the Breast or Breast Biomarker Reporting cancer protocols
published by the College of American Pathologists for the AJCC Staging System Breast

202 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3827: Estrogen Receptor Summary
Item Length: 1
NAACCR Item #: 3827
XML Parent-NAACCR ID: Tumor-estrogenReceptorSummary
NAACCR Alternate Name: ER (Estrogen Receptor) Summary
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Estrogen Receptor Summary is a summary of results of the estrogen receptor (ER) assay.
Rationale
This data item is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 48, Breast. It was
previously collected as Breast CS SSF # 1.
Coding guidelines
Record the pathologist’s interpretation of the assay value from the tumor specimen. Results from the
ER assay done prior to neoadjuvant therapy take priority. If there are no results prior to neoadjuvant
treatment, code the results from a post-treatment specimen. Do not report the results of an ER or PR
done as part of a multigene test such as OncotypeDX or MammaPrint.
Code 0 when the ER is reported as negative or normal
Code 1 when the ER is reported as positive or elevated
Code 7 when the ER test was ordered but the results are not available
Code 9 when the ER is
o Reported as borderline; undetermined whether positive or negative
o Cannot be determined by the pathologist (e.g. inadequate specimen)
o It is unknown whether the ER test was performed
o The patient has only a clinical diagnosis of breast cancer (no tissue diagnosis)
See 3826-3827, 3914-3916: Estrogen Receptor and Progesterone Receptor for additional information.
Coding Instructions and Codes
Note 1: Physician statement of ER (Estrogen Receptor) Summary status can be used to code this data
item when no other information is available.
Note 2: The result of the ER test performed on the primary breast tissue is to be recorded in this data
item.
Note 3: Results from nodal or metastatic tissue may be used ONLY when there is no evidence of in situ
or invasive carcinoma in the primary tumor.
Note 4: In cases where there are invasive and in situ components in the primary tumor and ER is done
on both, ignore the in situ results.

203 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
If ER is positive on an in situ component and ER is negative on all tested invasive components in
the primary tumor, code ER as negative (code 0)
If in situ and invasive components present and ER only done on the in situ component in the
primary tumor, code unknown (code 9)
Note 5: In cases where there is a single tumor with multiple biopsies and/or surgical resection with
different ER results.
Use the highest (positive versus negative)
Note 6: In cases where there are multiple tumors with different ER results, code the results from the
largest tumor size (determined either clinically or pathologically) when multiple tumors are present.
Do not use specimen size to determine the largest tumor size
Note 7: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy. If neoadjuvant therapy is given and there are no ER results from pre-treatment specimens,
report the findings from post-treatment specimens.
Note 8: If the patient is ER positive and node negative, a multigene test such as Oncotype Dx may be
performed, in which case another ER test will be performed. Do not record the results of that test in
this field.
Record only the results of the test which made the patient eligible to be given the multigene test
Code Description
0 ER negative (0.0% or less than 1%)
1 ER positive
7
Test ordered, results not in chart
9 Not documented in medical record
Cannot be determined (indeterminate)
ER (Estrogen Receptor) Summary status not assessed or unknown if
assessed
Return to Schema ID Table

204 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3826: Estrogen Receptor Percent Positive or Range
Item Length: 3
NAACCR Item #: 3826
XML Parent-NAACCR ID: Tumor-estrogenReceptorPercntPosOrRange
NAACCR Alternate Name: ER (Estrogen Receptor) Percent Positive or Range
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Estrogen Receptor Percent Positive or Range is the percent of cells staining estrogen receptor positive
by IHC.
Rationale
Estrogen Receptor Percent Positive or Range is a Registry Data Collection Variable in AJCC. It is a new
data item for cases diagnosed 1/1/2018+.
See 3826-3827, 3914-3916: Estrogen Receptor and Progesterone Receptor for additional information.
Coding Instructions and Codes
Note 1: Physician statement of ER Percent Positive or Range can be used to code this data item.
Note 2: Code this data item using the same report used to record 3827: Estrogen Receptor Summary.
Note 3: If ER is negative, or percentage is less than 1%, code 000.
Note 4: The actual ER (1-100%) percent takes priority over the range codes.
Note 5: If ER is positive but percentage is unknown, code XX7.
Note 6: Ranges for the codes in this data item are defined in steps of 10 which correspond to the CAP
protocol. If a range in a report is given in steps other than those provided in the codes, code to the
range that contains the lowest number of the range in the report.
Example 1: Report says 1-5%. Code R10 (1-10%)
Example 2: Report says 90-95%. Code R90 (81-90%)
Code Description
000 ER negative, or stated as less than 1%
001-100 1-100 percent
R10 Stated as 1-10%
R20 Stated as 11-20%
R30 Stated as 21-30%
R40 Stated as 31-40%
R50 Stated as 41-50%
R60 Stated as 51-60%
R70 Stated as 61-70%

205 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
R80 Stated as 71-80%
R90 Stated as 81-90%
R99 Stated as 91-100%
XX7 Test done, results not in chart
XX8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XX8 will result in an edit error.)
XX9
Not documented in medical record
ER (Estrogen Receptor) Percent Positive or Range not assessed or unknown if assessed
Return to Schema ID Table

206 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3828: Estrogen Receptor Total Allred Score
Item Length: 2
NAACCR Item #: 3828
XML Parent-NAACCR ID: Tumor-estrogenReceptorTotalAllredScore
NAACCR Alternate Name: ER (Estrogen Receptor) Total Allred Score
Active years: 2018-2022
Schemas(s):
00480: Breast (2018+)
Description
Estrogen Receptor Total Allred Score is based on the percentage of cells that stain positive by IHC for
estrogen receptor (ER) and the intensity of that staining.
Rationale
Estrogen Receptor Total Allred Score is a Registry Data Collection Variable in AJCC. It is a new data item
for cases diagnosed 1/1/2018+.
See 3826-3827, 3914-3916: Estrogen Receptor and Progesterone Receptor for additional information.
Coding Instructions and Codes
Note 1: This SSDI is no longer required by any of the standard setters starting with 2023 diagnoses.
For cases diagnosed 2023+, this SSDI may be left blank
Note 2: Physician statement of ER (Estrogen Receptor) Total Allred Score can be used to code this data
item.
Note 3: Code this data item using the same report used to record 3827: Estrogen Receptor Summary.
Note 4: The Allred system looks at what percentage of cells test positive for hormone receptors, along
with how well the receptors show up after staining (this is called “intensity”). This information is then
combined to score the sample on a scale from 0 to 8. The higher the score, the more receptors were
found and the easier they were to see in the sample.
The registrar should not calculate the Allred score unless both components are available
(proportion score and intensity)
See the Allred Score for Estrogen and Progesterone Receptor Evaluation section in the SSDI
manual for assistance in determining the Allred Score
Note 5: If ER test is performed, but Allred score is not documented, or cannot be calculated, code X9.
Code Description
00 Total ER Allred score of 0
01 Total ER Allred score of 1
02 Total ER Allred score of 2
03 Total ER Allred score of 3
04 Total ER Allred score of 4

207 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
05 Total ER Allred score of 5
06 Total ER Allred score of 6
07 Total ER Allred score of 7
08 Total ER Allred score of 8
X8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9
Not documented in medical
record
ER (Estrogen Receptor) Total Allred Score not assessed, or unknown if assessed
<Blank> NA-Diagnosis years is after 2022
Return to Schema ID Table

208 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3915: Progesterone Receptor Summary
Item Length: 1
NAACCR Item #: 3915
XML Parent-NAACCR ID: Tumor-progesteroneRecepSummary
NAACCR Alternate Name: PR (Progesterone Receptor) Summary
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Progesterone Receptor Summary is a summary of results from the progesterone receptor (PR) assay.
Rationale
This data item is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 48 Breast. It was
previously collected as Breast CS SSF # 2.
Coding guidelines
Code 0 when the PR is reported as negative or normal
Code 1 when the PR is reported as positive or elevated
Code 7 when the PR test was ordered but the results are not available
Code 9 when the PR is
o Reported as borderline; undetermined whether positive or negative
o Cannot be determined by the pathologist (e.g. inadequate specimen)
o It is unknown whether the PR test was performed
o The patient has only a clinical diagnosis of breast cancer (no tissue diagnosis)
See 3826-3827, 3914-3916: Estrogen Receptor and Progesterone Receptor for additional information.
Coding Instructions and Codes
Note 1: Physician statement of PR (Progesterone Receptor) Summary status can be used to code this
data item when no other information is available.
Note 2: The result of the PR test performed on the primary breast tissue is to be recorded in this data
item.
Note 3: Results from nodal or metastatic tissue may be used ONLY when there is no evidence of in situ
or invasive carcinoma in the primary tumor.
Note 4: In cases where there are invasive and in situ components in the primary tumor and PR is done
on both, ignore the in situ results.
If PR is positive on an in situ component and PR is negative on all tested invasive components in
the primary tumor, code PR as negative (code 0)

209 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
If in situ and invasive components present and PR only done on the in situ component in the
primary tumor, code unknown (code 9)
Note 5: In cases where there is a single tumor with multiple biopsies and/or surgical resection with
different PR results.
Use the highest (positive versus negative)
Note 6: In cases where there are multiple tumors with different PR results, code the results from the
largest tumor size (determined either clinically or pathologically) when multiple tumors are present.
Do not use specimen size to determine the largest tumor size
Note 7: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy. If neoadjuvant therapy is given and there are no PR results from pre-treatment specimens,
report the findings from post-treatment specimens.
Note 8: If the patient is PR positive and node negative, a multigene test such as Oncotype Dx may be
performed, in which case another PR test will be performed. Do not record the results of that test in
this field.
Record only the results of the test which made the patient eligible to be given the multigene test
Code Description
0 PR negative (0.0% or less than 1%)
1 PR positive
7 Test ordered, results not in chart
9 Not documented in medical record
Cannot be determined (indeterminate)
PR (Progesterone Receptor) Summary status not assessed or
unknown if assessed
Return to Schema ID Table

210 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3914: Progesterone Receptor Percent Positive or Range
Item Length: 3
NAACCR Item #: 3914
XML Parent-NAACCR ID: Tumor-progesteroneRecepPrcntPosOrRange
NAACCR Alternate Name: PR (Progesterone Receptor) Percent Positive or Range
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Progesterone Receptor Percent Positive or Range is the percent of cells staining progesterone receptor
positive measured by IHC.
Rationale
Progesterone Receptor Percent Positive or Range is a Registry Data Collection Variable in AJCC. It is a
new data item for cases diagnosed 1/1/2018+.
See 3826-3827, 3914-3916: Estrogen Receptor and Progesterone Receptor for additional information.
Coding Instructions and Codes
Note 1: Physician statement of PR (Progesterone Receptor) Percent Positive or Range can be used to
code this data item.
Note 2: Code this data item using the same report used to record 3915: Progesterone Receptor
Summary.
Note 3: If PR is negative, or percentage is less than 1%, code 000.
Note 4: The actual PR (1-100%) percent takes priority over the range codes.
Note 5: If PR is positive but percentage is unknown, code XX7.
Note 6: Ranges for the codes in this data item are defined in steps of 10 which correspond to the CAP
protocol. If a range in a report is given in steps other than those provided in the codes, code to the
range that contains the lowest number of the range in the report.
Example 1: Report says 1-5%. Code R10 (1-10%)
Example 2: Report says 90-95%. Code R90 (81-90%)
Code Description
000
PR negative, or stated as less than 1%
001-100 1-100 percent
R10 Stated as 1-10%
R20 Stated as 11-20%
R30 Stated as 21-30%
R40 Stated as 31-40%

211 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
R50 Stated as 41-50%
R60 Stated as 51-60%
R70 Stated as 61-70%
R80 Stated as 71-80%
R90 Stated as 81-90%
R99
Stated as 91
-
100%
XX7 Test done, results not in chart
XX8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XX8 will result in an edit
error.)
XX9
Not documented in medical record
PR (Progesterone Receptor) Percent Positive or Range not assessed or unknown if
assessed
Return to Schema ID Table

212 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3916: Progesterone Receptor Total Allred Score
Item Length: 2
NAACCR Item #: 3916
XML Parent-NAACCR ID: Tumor-progesteroneRecepTotalAllredScor
NAACCR Alternate Name: PR (Progesterone Receptor) Total Allred Score
Active years: 2018-2022
Schemas(s):
00480: Breast (2018+)
Description
Progesterone Receptor, Total Allred Score is based on the percentage of cells that stain by IHC for
progesterone receptor (PR) and the intensity of that staining.
Rationale
Progesterone Receptor, Total Allred Score is a Registry Data Collection Variable in AJCC. It is a new data
item for cases diagnosed 1/1/2018+.
See 3826-3827, 3914-3916: Estrogen Receptor and Progesterone Receptor for additional information.
Coding Instructions and Codes
Note 1: This SSDI is no longer required by any of the standard setters starting with 2023 diagnoses.
For cases diagnosed 2023+, this SSDI may be left blank
Note 2: Physician statement of PR (Progesterone Receptor) Total Allred Score can be used to code this
data item.
Note 3: Code this data item using the same report used to record 3915: Progesterone Receptor
Summary.
Note 4: The Allred system looks at what percentage of cells test positive for hormone receptors, along
with how well the receptors show up after staining (this is called “intensity”). This information is then
combined to score the sample on a scale from 0 to 8. The higher the score, the more receptors were
found and the easier they were to see in the sample.
The registrar should not calculate the Allred score unless both components are available
(proportion score and intensity)
See the Allred Score for Estrogen and Progesterone Receptor Evaluation section in the SSDI
manual for assistance in determining the Allred Score
Note 5: If PR test is performed, but Allred score is not documented, or it cannot be calculated, code X9.
Code
Description
00
Total PR Allred score of 0
01 Total PR Allred score of 1
02 Total PR Allred score of 2
03 Total PR Allred score of 3

213 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
04 Total PR Allred score of 4
05 Total PR Allred score of 5
06 Total PR Allred score of 6
07 Total PR Allred score of 7
08 Total PR Allred score of 8
X8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit
error.)
X9 Not documented in medical record
PR (Progesterone Receptor) Total Allred Score not assessed, or unknown if
assessed
<Blank>
NA
-
Diagnosis years is after 2022
Return to Schema ID Table

214 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
HER2
Definition
A subset of breast carcinomas (approximately 15% to 20%) overexpress human epidermal growth factor
receptor 2 (HER2). The presence of HER2 overexpression in untreated patients is associated with worse
prognosis in both node-negative and node-positive patients. Protein overexpression is usually due to
HER2 gene amplification. The HER2 protein may also be referred to as ERBB2 and the HER2 gene may
also be referred to as the ERBB2 gene.
The development of HER-2 targeting agents for the treatment of HER2 positive breast cancer has
dramatically improved outcomes for patients with HER2 positive breast cancers. HER2 status is
primarily evaluated to determine patient eligibility for anti-HER2 therapy.
The following data items are used to collect HER2 information:
3850: HER2 IHC Summary
3851: HER2 ISH Dual Probe Copy Number
3852: HER2 ISH Dual Probe Ratio
3853: HER2 ISH Single Probe Copy Number
3854: HER2 ISH Summary
3855: HER2 Overall Summary
The simplest test used is the IHC (immunohistochemistry). If the IHC test is borderline or indeterminate,
an ISH (in situ hybridization) test may be performed.
The results of the IHC test are reported as follows:
Reporting Results of HER2 Testing by Immunohistochemistry (IHC)
Result
Criteria
Negative (Score 0)
No staining observed
or
tumor cells
Negative (Score 1+) Incomplete, faint/barely perceptible membrane staining in >10% of invasive
tumor cells*
Equivocal (Score 2+)
Incomplete and/or weak to moderate circumferential membrane staining in
>10% of invasive tumor cells
or
tumor cells*
Positive (Score 3+) Complete, intense, circumferential membrane staining in >10% of invasive
tumor cells*
If the IHC test is borderline or indeterminate, an ISH test may be performed. The ISH test is a method of
testing for overexpression of the HER2 gene that uses fluorescent pieces of DNA that attach only to the

215 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
HER2 gene copies in cells, which can then be counted under a special microscope. ISH studies
determine the presence or absence of gene amplification and methods include fluorescence in situ
hybridization (FISH), chromogenic in situ hybridization (CISH), and silver-enhanced in situ hybridization
(SISH). Some assays use a single probe to determine the number of HER2 gene copies present (single-
probe assays) and others include a chromosome enumeration probe (CEP17) to determine the ratio of
HER2 signals to copies of chromosome 17 (dual-probe assays).
Results from single probe and dual probe ISH tests are reported differently and are collected in different
data items. For dual probe tests, both HER2/CEP17 ratio and HER2 copy number results are collected in
separate data items.
Reporting Results of HER2 Testing by In Situ Hybridization (single-probe assay)
Result Criteria
Negative (not
amplified)
Average HER2 copy number <4.0 signals/cell
Equivocal Average HER2 copy nu
Positive (amplified) Average HER2
Reporting Results of HER2 Testing by In Situ Hybridization (dual-probe assay)
Result Criteria
Negative (not
amplified)
HER2/CEP17 ratio <2.0 AND average HER2 copy number <4.0 signals/cell
Equivocal HER2/CEP17
signals/cell
Positive (amplified) HER2/CEP17 (regardless of average HER2 copy number)
or
Average HER (regardless of ratio)
Note: TP52, SMSCR and RARA are gene genes that are also on chromosome 17. However, they are not
close to the centromere, and thus can be used to assess borderline/equivocal fish results (ratios) when
the centromeric probe for chromosome 17 (CEP17) performance may be problematic. Although these
may be helpful in some cases, they are not the same as the CEP17 result or the ratio determined from
CEP17. There should always be a prior CEP17 result when these other results are found in the chart. If
one of these tests (TP52, SMSCR, RARA, or others) are used and a dual probe copy number/ratio are
documented, record that results in the appropriate data item.
D17Z1 is the CEP17 probe used in the Vysis (Abbot) FISH kit. So, for the HER2 data items, D17Z1 and
CEP17 are to be treated as the same thing.
Changes from Collaborative Stage v2 (CSv2): In CSv2, there were multiple SSFs that collected
information on FISH, CISH, or other. In addition, the lab value and the interpretation were collected. For
2018 cases forward, only the interpretation will be recorded. Also, interpretation of all types of ISH tests
(FISH, CISH, SISH, single probe, double probe) are to be recorded in the overall ISH data item. If there are
multiple tests, record the highest.

216 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note: HER2 results are to be recorded from IHC or ISH tests only. Do not use results from the following
tests to record HER2 results
Oncotype Dx
MammaPrint
EndoPredict
PAM 50 (Prosigna)
Any other test that records HER2
Additional Information
For further information, refer to the Breast or Breast Biomarker Reporting cancer protocols
published by the College of American Pathologists for the AJCC Staging System Breast

217 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3850: HER2 IHC Summary
Item Length: 1
NAACCR Item #: 3850
XML Parent-NAACCR ID: Tumor-her2IhcSummary
NAACCR Alternate Name: None
Active years: 2018-2020
Schemas(s):
00480: Breast (2018+)
Description
HER2 IHC Summary is the summary score for HER2 testing by IHC.
Rationale
HER2 IHC Summary is a Registry Data Collection Variable in AJCC. It is a new data item for cases
diagnosed 1/1/2018+.
See 3850-3855: HER2 for additional information.
Coding Instructions and Codes
Note 1: This SSDI is no longer required by any of the standard setters starting with 2021 diagnoses.
For cases diagnosed 2021+, this SSDI may be left blank
Note 2: Physician statement of HER2 IHC Summary can be used to code this data item when no other
information is available.
Note 3: The HER2 IHC test performed on the primary breast tissue is to be recorded in this data item.
Note 4: Results from nodal or metastatic tissue may be used, ONLY when there is no evidence of
primary tumor.
Note 5: In cases where there are invasive and in situ components and HER2 IHC is done on both, ignore
the in situ results.
If HER2 IHC is positive on an in situ component and HER2 IHC is negative on all tested invasive
components, code HER2 IHC as negative (code 0)
If in situ and invasive components present and HER2 IHC only done on the in situ component,
code unknown (code 9)
Note 6: In cases where there is a single tumor with multiple biopsies and/or surgical resection with
different HER2 IHC results.
Use the highest (positive versus negative)
Note 7: In cases where there are multiple tumors with different HER2 IHC results, code the results from
the largest tumor size (determined either clinically or pathologically) when multiple tumors are present.

218 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Do not use specimen size to determine the largest tumor size
Note 8: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no HER2 IHC results from pre-treatment
specimens, report the findings from post-treatment specimens.
Note 9: A 2+ (equivocal) finding by IHC should result in additional testing with ISH to determine gene
copy number.
Note 10: An immunohistochemistry (IHC) test identifies the protein expressed by the gene (ERBB2), and
an in situ hybridization (ISH) test identifies the number of copies of the gene (ERBB2) itself.
Note 11: HER2 is not routinely done on pure in situ tumors (behavior /2); however, if you have an in situ
tumor and there are HER2 results, go ahead and record it. Otherwise code 9.
Code Description
0 Negative (Score 0)
1 Negative (Score 1+)
2 Equivocal (Score 2+)
Stated as equivocal
Borderline
3
Positive (Score 3+)
Stated as positive
4 Stated as negative, but score not stated
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Cannot be determined (indeterminate)
HER2 IHC Summary not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is after 2020
Return to Schema ID Table

219 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3854: HER2 ISH Summary
Item Length: 1
NAACCR Item #: 3854
XML Parent-NAACCR ID: Tumor-her2IshSummary
NAACCR Alternate Name: None
Active years: 2018-2020
Schemas(s):
00480: Breast (2018+)
Description
HER2 in situ hybridization (ISH) Summary is the summary score for results of testing for ERBB2 gene
copy number by any ISH method. An immunohistochemistry (IHC) test identifies the protein expressed
by the gene (ERBB2), and an ISH test identifies the number of copies of the gene (ERBB2) itself.
Rationale
HER2 ISH Summary is a Registry Data Collection Variable in AJCC. It is a new data item for cases
diagnosed 1/1/2018+.
See 3850-3855: HER2 for additional information.
Coding Instructions and Codes
Note 1: This SSDI is n o longer required by any of the standard setters starting with 2021 diagnoses.
For cases diagnosed 2021+, this SSDI may be left blank
Note 2: Physician statement of HER2 in situ hybridization (ISH) Summary can be used to code this data
item when no other information is available.
Note 3: The HER2 ISH test performed on the primary breast tissue is to be recorded in this data item.
Note 4: Results from nodal or metastatic tissue may be used, ONLY when there is no evidence of
primary tumor.
Note 5: Any type of ISH test (e.g., FISH, CISH, SISH) can be used to code this data item. The same test
should be used to code all the HER2 ISH data items.
Note 6: In cases where there are invasive and in situ components and HER2 ISH is done on both, ignore
the in situ results.
If HER2 ISH is positive on an in situ component and HER2 ISH is negative on all tested invasive
components, code HER2 ISH as negative (code 0)
If in situ and invasive components present and HER2 ISH only done on the in situ component,
code unknown (code 9)
Note 7: In cases where there is a single tumor with multiple biopsies and/or surgical resection with
different HER2 ISH results.

220 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Use the highest (positive versus negative)
Note 8: In cases where there are multiple tumors with different HER2 ISH results, code the results from
the largest tumor size (determined either clinically or pathologically) when multiple tumors are present.
Do not use specimen size to determine the largest tumor size
Note 9: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no HER2 ISH results from pre-treatment
specimens, report the findings from post-treatment specimens.
Note 10: An immunohistochemistry (IHC) test identifies the protein expressed by the gene (ERBB2), and
an ISH test identifies the number of copies of the gene (ERBB2) itself.
Note 11: HER2 is not routinely done on pure in situ tumors (behavior /2); however, if you have an in situ
tumor and there are HER2 results, go ahead and record it. Otherwise code 9.
Code Description
0 Negative [not amplified]
2 Equivocal
3 Positive [amplified]
7 Test ordered, results not in chart
8
Not applicable: Information not
collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit
error.)
9 Not documented in medical record
Results cannot be determined (indeterminate)
Borderline
HER2 ISH Summary not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is after 2020
Return to Schema ID Table

221 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3855: HER2 Overall Summary
Item Length: 1
NAACCR Item #: 3855
XML Parent-NAACCR ID: Tumor-her2OverallSummary
NAACCR Alternate Name: None
Active years: 2018+Schemas(s):
00480: Breast (2018+)
Description
HER2 Overall Summary is a summary of results from HER2 testing.
Rationale
This data item is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 48 Breast. It was
previously collected as Breast, CS SSF # 15. Experts recommend that every invasive breast cancer be
tested for the presence of HER2 because anti-HER2 treatments are highly effective for these tumors.
HER2 overall summary It will be collected for Esophagus and Esophagogastric Junction and Stomach for
cases diagnosed 1/1/21+ because NCCN guidelines recommend HER2 testing at time of diagnosis if
patients are documented or suspected of having metastatic disease. HER2 monoclonal antibodies may
be added to chemotherapy for patients with HER2 positive disease.
See 3850-3855: HER2 for additional information.
Coding guidelines
Record the pathologist’s interpretation of the HER2 test from the tumor specimen. Results from the
HER2 test done prior to neoadjuvant therapy take priority. If there are no results prior to neoadjuvant
treatment, code the results from a post-treatment specimen. Do not report the results of a HER2 as
part of a multigene test such as OncotypeDX or MammaPrint.
If assays are performed on more than one specimen and any result is interpreted as positive, code as 1
Positive/elevated.
Exception: If results from both an in situ specimen and an invasive component are given, record the
results from the invasive specimen, even if the in situ is positive and the invasive specimen is negative.
Code 0 when the HER2 is reported as negative or normal
Code 1 when the HER2 is reported as positive or elevated
Code 7 when the HER2 test was ordered but the results are not available
Code 9 when the HER2 is
o Reported as borderline; undetermined whether positive or negative
o Cannot be determined by the pathologist (e.g. inadequate specimen)
o It is unknown whether the HER2 test was performed
o The patient has only a clinical diagnosis of breast cancer (no tissue diagnosis)

222 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note 1: Physician statement of HER2 Overall Summary can be used to code this data item when no
other information is available.
Note 2: The result of the HER2 test performed on the primary breast tissue is to be recorded in this data
item.
Note 3: Results from nodal or metastatic tissue may be used ONLY when there is no evidence of in situ
or invasive carcinoma in the primary tumor.
Note 4: In cases where there are invasive and in situ components in the primary tumor and HER2 is done
on both, ignore the in situ results.
If HER2 is positive on an in situ component and HER2 is negative on all tested invasive
components in the primary tumor, code HER2 as negative (code 0)
If in situ and invasive components present and HER2 only done on the in situ component in the
primary tumor, code unknown (code 9)
Note 5: In cases where there is a single tumor with multiple biopsies and/or surgical resection with
different HER2 results.
Use the highest (positive versus negative)
Note 6: In cases where there are multiple tumors with different HER2 results, code the results from the
largest tumor size (determined either clinically or pathologically) when multiple tumors are present.
Do not use specimen size to determine the largest tumor size
Note 7: If neoadjuvant therapy is given, record the assay from tumor specimens prior to neoadjuvant
therapy.
If neoadjuvant therapy is given and there are no HER2 results from pre-treatment specimens,
report the findings from post-treatment specimens
Note 8: If the patient is HER2 positive and node negative, a multigene test such as Oncotype Dx may be
performed, in which case another HER2 test will be performed. Do not record the results of that test in
this field.
Record only the results of the test which made the patient eligible to be given the multigene test
Note 9: HER2 is not routinely done on pure in situ tumors (behavior /2); however, if you have an in situ
tumor and there are HER2 results, go ahead and record it. Otherwise code 9
Code Description
0 HER2 negative; equivocal
1 HER2 positive
7 Test ordered, results not in chart
9 Not documented in medical record
Cannot be determined (indeterminate)
Borderline
HER2 Overall Summary status not assessed or unknown if assessed
Return to Schema ID Table

223 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3853: HER2 ISH Single Probe Copy Number
Item Length: 4
NAACCR Item #: 3853
XML Parent-NAACCR ID: Tumor-her2IshSingleProbeCopyNumber
NAACCR Alternate Name: None
Active years: 2018-2020
Schemas(s):
00480: Breast (2018+)
Description
HER2 in situ hybridization (ISH) Single Probe Copy Number is the HER2 copy number based on a single
probe test.
Rationale
HER2 ISH Single Probe Copy Number is a Registry Data Collection Variable in AJCC. It is a new data item
for cases diagnosed 1/1/2018+.
See 3850-3855: HER2 for additional information.
Coding Instructions and Codes
Note 1: This SSDI is no longer required by any of the standard setters starting with 2021 diagnoses.
For cases diagnosed 2021+, this SSDI may be left blank
Note 2: Physician statement of HER2 in situ hybridization (ISH) Single Probe Copy Number can be used
to code this data item.
Note 3: A single probe test will report average number or mean signals per cell for HER2. Record the
HER2 average number or mean signals per cells in this data item. The average number or mean signals
per cell is also called the copy number.
Example:
SISH RESULTS: FINAL HER 2 IN SITU HYBRIDIZATION INTERPRETATION: POSITIVE (>6 gene copies)
HER-2/neu gene amplification.
HER-2/neu SILVER IN SITU HYBRIDIZATION (SISH)
HER-2neu gene (Inform HER2 DNA probe)
Number of tumor cell nuclei counted: 60
Number of Her-2/neu gene copies: 418
Mean HER-2/neu gene copy number: 6.9
Code Single Probe HER2 Copy Number: 6.9

224 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
[Note: This is calculated by dividing 418 by 60]
Note 4: Registrars are not to calculate the copy number.
Note 5: Following ASCO-CAP guidelines, a 2+ (equivocal) finding by immunohistochemistry (IHC) should
result in additional testing with ISH to determine gene copy number.
Note 6: Any type of ISH test (e.g., FISH, CISH, SISH) can be used to code this data item. Code this data
item using the same report used to record 3854: HER2 ISH Summary.
Note 7: A HER2 ISH test may be called “ERBB2.” ERBB2 is the standard symbol for the gene ‘erb-b2
receptor tyrosine kinase 2.’ An IHC test identifies the protein expressed by the gene, and an ISH test
identifies the gene itself.
Note 8: If a HER2 ISH single probe copy number test is done, and the results are between 4 and 6
(equivocal), dual probe tests are recommended.
Note 9: If the test results are presented to the hundredth decimal, ignore the hundredth decimal. Do
NOT round.
Example:
Reported as 6.97, code 6.9
Code Description
0.0-99.9 Reported HER2 copy number of 0.0-99.9
XX.1 Reported HER2 copy number of 100 or greater
XX.7 Test ordered, results not in chart
XX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XX.8 will result in an edit
error.)
XX.9 Not documented in medical record
Cannot be determined (indeterminate)
Single probe test not done; only dual probe test performed
HER2 ISH Single Probe Copy Number not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is after 2020
Return to Schema ID Table

225 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3851: HER2 ISH Dual Probe Copy Number
Item Length: 4
NAACCR Item #: 3851
XML Parent-NAACCR ID: Tumor-her2IshDualProbeCopyNumber
NAACCR Alternate Name: None
Active years: 2018-2020
Schemas(s):
00480: Breast (2018+)
Description
HER2 in situ hybridization (ISH) Dual Probe Copy Number is the HER2 copy number based on a dual
probe test.
Rationale
HER2 ISH Dual Probe Copy Number is a Registry Data Collection Variable in AJCC. It is a new data item
for cases diagnosed 1/1/2018+.
See 3850-3855: HER2 for additional information.
Coding Instructions and Codes
Note 1: This SSDI is no longer required by any of the standard setters starting with 2021 diagnoses.
For cases diagnosed 2021+, this SSDI may be left blank
Note 2: Physician statement of HER2 in situ hybridization (ISH) Dual Probe Copy Number can be used to
code this data item.
Note 3: A dual probe test will report average number or mean signals per cell for both HER2 and CEP17,
the latter used as a control. Record the HER2 average number or mean signals per cells in this data item.
The average number or mean signals per cells is also called the copy number.
Example:
SISH RESULTS: FINAL HER 2 IN SITU HYBRIDIZATION INTERPRETATION: EQUIVOCAL,
INDETERMINATE. HER2 gene copy between 4 & 6 with HER2/CEP17 ratio <2.
HER2/CEP17 RATIO: 4.26 / 3.13 = 1.36
HER-2/neu SILVER IN SITU HYBRIDIZATION (SISH)
HER-2neu gene (Inform HER2 DNA probe)
Number of tumor cell nuclei counted: 120
Number of Her-2/neu gene copies: 511
Mean HER-2/neu gene copy number: 4.26

226 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
CEP-17 SILVER IN SITU HYBRIDIZATION (SISH)
CEP-17 (Inform Chromosome 17 probe)
Number of cell nuclei counted: 60
Number of CEP-17 gene copies: 188
Mean CEP-17 gene copies/nucl: 3.13
Code Dual Probe HER2 Copy Number: 4.2
[Note: This is calculated by dividing 511 by 120]
Note 4: Registrars are not to calculate the copy number.
Note 5: Following ASCO-CAP guidelines, a 2+ (equivocal) finding by immunohistochemistry (IHC) should
result in additional testing with ISH to determine gene copy number.
Note 6: Any type of ISH test (e.g., FISH, CISH, SISH) can be used to code this data item. Code this data
item using the same report used to record 3854: HER2 ISH Summary.
Note 7: A HER2 ISH test may be called “ERBB2.” ERBB2 is the standard symbol for the gene ‘erb-b2
receptor tyrosine kinase 2.’ An IHC test identifies the protein expressed by the gene, and an ISH test
identifies the gene itself.
Note 8: If a HER2 ISH single probe copy number test is done, and the results are between 4 and 6
(equivocal), dual probe tests are recommended.
Note 9: If the test results are presented to the hundredth decimal, ignore the hundredth decimal. Do
NOT round.
Example:
Reported as 4.99, code as 4.9
Code Description
0.0-99.9 Reported HER2 copy number of 0.0-99.9
XX.1 Reported HER2 copy number of 100 or greater
XX.7 Test ordered, results not in chart
XX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XX.8 will result in an edit
error.)
XX.9 Not documented in medical record
Cannot be determined (indeterminate)
Dual probe test note done; only single probe test performed
HER2 ISH Dual Probe Copy Number not assessed or unknown if assessed
<Blank>
N/A
-
Diagnosis year is
after 2020
Return to Schema ID Table

227 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3852: HER2 ISH Dual Probe Ratio
Item Length: 4
NAACCR Item #: 3852
XML Parent-NAACCR ID: Tumor-her2IshDualProbeRatio
NAACCR Alternate Name: None
Active years: 2018-2020
Schemas(s):
00480: Breast (2018+)
Description
HER2 ISH Dual Probe Ratio is the summary score for HER2 testing using a dual probe. The test will
report results for both HER2 and CEP17, the latter used as a control. The HER2/CEP17 ratio is reported.
Rationale
HER2 ISH Dual Probe Ratio is a Registry Data Collection Variable in AJCC. It is a new data item for cases
diagnosed 1/1/2018+.
See 3850-3855: HER2 for additional information.
Coding Instructions and Codes
Note 1: This SSDI is no longer required by any of the standard setters starting with 2021 diagnoses.
For cases diagnosed 2021+, this SSDI may be left blank
Note 2: Physician statement of HER2 in situ hybridization (ISH) Dual Probe Ratio can be used to code this
data item.
Note 3: A dual probe test will report results for both HER2 and CEP17, the latter used as a control. The
HER2/CEP17 ratio will be reported. Record the ratio in this data item.
Example:
SISH RESULTS: FINAL HER 2 IN SITU HYBRIDIZATION INTERPRETATION: EQUIVOCAL,
INDETERMINATE. HER2 gene copy between 4 & 6 with HER2/CEP17 ratio <2.
HER2/CEP17 RATIO: 4.26 / 3.13 = 1.36
HER-2/neu SILVER IN SITU HYBRIDIZATION (SISH)
HER-2neu gene (Inform HER2 DNA probe)
Number of tumor cell nuclei counted: 120
Number of Her-2/neu gene copies: 511
Mean HER-2/neu gene copy number: 4.26

228 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
CEP-17 SILVER IN SITU HYBRIDIZATION (SISH)
CEP-17 (Inform Chromosome 17 probe)
Number of cell nuclei counted: 60
Number of CEP-17 gene copies: 188
Mean CEP-17 gene copies/nucl: 3.13
Code Dual Probe HER2 Copy Number: 4.2
Code Dual Probe Ratio: 1.3
Note 4: Registrars are not to calculate the ratio.
Note 5: Following ASCO-CAP guidelines, a 2+ (equivocal) finding by immunohistochemistry (IHC) should
result in additional testing with ISH to determine gene copy number.
Note 6: Any type of ISH test (e.g., FISH, CISH, SISH) can be used to code this data item. Code this data
item using the same report used to record 3854: HER2 ISH Summary.
Note 7: A HER2 ISH test may be called “ERBB2.” ERBB2 is the standard symbol for the gene ‘erb-b2
receptor tyrosine kinase 2.’ An IHC test identifies the protein expressed by the gene, and an ISH test
identifies the gene itself.
Note 8: If the test results are presented to the hundredth decimal, ignore the hundredth decimal. Do
NOT round.
Example:
Reported as 1.99, code as 1.9
Code Description
0.0-99.9 Ratio of 0.0 to 99.9
XX.2 Less than 2.0
XX.3 Greater than or equal to 2.0
XX.7 Test ordered, results not in chart
XX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XX.8 will result in an
edit error.)
XX.9 Not documented in medical record
Results cannot be determined (indeterminate)
Dual probe test not done; only single probe test performed
HER2 ISH Dual Probe Ratio not assessed or unknown if assessed
<Blank> N/A-Diagnosis year is after 2020
Return to Schema ID Table

229 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
Multigene Signature Method and Results
Definition
Multigene testing is usually done for node-negative female breast cancer patients to predict risk of
recurrence within a specified time period or to predict the likelihood that the patient will respond to
specific types of chemotherapy. Multigene testing helps tailor treatment for the woman’s specific
cancer characteristics. Recent studies indicate that these tests may also be helpful in planning
treatment and predicting recurrence in node positive women with small tumors. Some types of tests
may be specific to ER positive or negative patients or women in a certain age range. Many different
types of genetic testing are available, including IHC-, FISH-, RT-PCR-, and genomic microarray-based
multigene predictors.
For the Breast cases, there are 2 data items that record information on Multigene testing.
3894: Multigene Signature Method
3895: Multigene Signature Results
These two fields record the type of multigene signature test that was performed. Both fields should be
coded from the same test, which may not be available at the time of diagnosis
Note: In Collaborative Stage v2 (CSv2), Oncotype was included in these two data items.
Oncotype has now been moved to separate data items. See the “Oncotype Dx” section of this
manual for more information.
Information is collected on the following tests
MammaPrint: A genomic test that analyzes the activity of certain genes in early-stage breast
cancer. Developed to help make treatment decisions based on the cancer's risk of coming back
(recurrence) within 10 years after diagnosis.
PAM 50 (Prosigna): PAM50 stands for Prediction Analysis of Microarray 50. It tests a sample of
the tumor (removed during a biopsy or surgery) for a group of 50 genes. Along with other
factors, the results of the PAM50 (Prosigna) test help predict the chance of metastasis (when
cancer spreads to other organs). Prosigna also helps to determine the molecular subtype of
breast cancer.
Breast Cancer Index: Analyzes the activity of seven genes to help predict the risk of node-
negative, hormone-receptor-positive breast cancer coming back 5 to 10 years after diagnosis.
The test can help women and their doctors decide if extending hormonal therapy 5 more years
(for a total of 10 years of hormonal therapy) would be beneficial. The Breast Cancer Index
reports two scores: how likely the cancer is to recur 5 to 10 years after diagnosis and how likely
a woman is to benefit from taking hormonal therapy for a total of 10 years.
EndoPredict: A genomic test for people newly diagnosed with early-stage, estrogen-receptor-
positive, HER2-negative breast cancer. May be used to help make treatment decisions based on
the cancer's risk of coming back in a part of the body away from the breast (distant metastasis)
within 10 years after diagnosis. The EndoPredict test provides a risk score that is either low-risk
or high-risk of breast cancer recurring as distant metastasis. Knowing if the cancer has a high or
low risk of recurrence can help women and their doctors decide if chemotherapy or other
treatments to reduce risk after surgery are needed.

230 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Source documents: specialty reference laboratories (private companies with proprietary testing
methods); the actual report may be included in the medical record or may be referenced by the
clinician.
Other names: genomic profiling, multigene testing, multigene assay, microarray assay,
molecular diagnostics for treatment planning
Return to Schema ID Table

231 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3894: Multigene Signature Method
Item Length: 1
NAACCR Item #: 3894
XML Parent-NAACCR ID: Tumor-multigeneSignatureMethod
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Multigene signatures or classifiers are assays of a panel of genes from a tumor specimen, intended to
provide a quantitative assessment of the likelihood of response to chemotherapy and to evaluate
prognosis or the likelihood of future metastasis. This data item identifies the multigene signature
method used. Oncotype Dx is coded elsewhere.
Rationale
Multigene Signature Method is a Registry Data Collection Variable in AJCC. It was previously collected as
Breast, CS SSF #22. See also Multigene Signature Results.
See 3894, 3895: Multigene Signature Method and Results for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the Multigene Signature Method can be used to code this data item.
Note 2: Multigene signatures or classifiers are assays of a panel of genes from a tumor specimen,
intended to provide a quantitative assessment of the likelihood of response to chemotherapy and to
evaluate prognosis or the likelihood of future metastasis.
Only record tests done on tumor tissue that help determine if the cancer is likely to recur. Don’t
include other tests, such as those that evaluate hereditary mutations that influence a patient’s
risk of developing cancer (e.g. myRisk, BRCA)
Only record tests that are based on gene assays. Don’t include other tests which use a
multivariate data model to eliminate the need for genetic assays
Note 3: Code the type of test performed. The same test should be used to record information in 3895:
Multigene Signature Results .
Note 4: Oncotype Dx tests are not recorded in this data item. See the following data items for Oncotype
Dx.
3903: Oncotype Dx Recurrence Score-DCIS
3904: Oncotype Dx Recurrence Score-Invasive
3905: Oncotype Dx Risk Level-DCIS
3906: Oncotype Dx Risk Level-Invasive

232 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
1 Mammaprint
2 PAM50 (Prosigna)
3 Breast Cancer Index
4 EndoPredict
5 Test performed, type of test unknown
6
Multiple tests, any tests in codes 1
-
4
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Multigene Signature Method not assessed or unknown if assessed
Return to Schema ID Table
233 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3895: Multigene Signature Results
Item Length: 2
NAACCR Item #: 3895
XML Parent-NAACCR ID: Tumor-multigeneSignatureResults
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Multigene signatures or classifiers are assays of a panel of genes from a tumor specimen, intended to
provide a quantitative assessment of the likelihood of response to chemotherapy and to evaluate
prognosis or the likelihood of future metastasis. This data item identifies the multigene signature result.
Oncotype Dx is coded elsewhere.
Rationale
Multigene Signature Results is a Registry Data Collection Variable in AJCC. It was previously collected as
Breast, CS SSF #23. See also Multigene Signature Method.
See 3894, 3895: Multigene Signature Method and Results for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the Multigene Signature Results can be used to code this data item.
Note 2: Multigene signatures or classifiers are assays of a panel of genes from a tumor specimen,
intended to provide a quantitative assessment of the likelihood of response to chemotherapy and to
evaluate prognosis or the likelihood of future metastasis.
Only record tests done on tumor tissue that help determine if the cancer is likely to recur. Don’t
include other tests, such as those that evaluate hereditary mutations that influence a patient’s
risk of developing cancer (e.g. myRisk, BRCA)
Only record tests that are based on gene assays. Don’t include other tests which use a
multivariate data model to eliminate the need for genetic assays
Note 3: Code the score or risk for the test performed. The same test should be used to record
information in 3894: Multigene Signature Method.
Note 4: Oncotype Dx tests are not recorded in this data item. See the following data items for Oncotype
Dx.
3903: Oncotype Dx Recurrence Score-DCIS
3904: Oncotype Dx Recurrence Score-Invasive
3905: Oncotype Dx Risk Level-DCIS
3906: Oncotype Dx Risk Level-Invasive

234 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 5: PAM50 (Prosigna) is a single numeric score of 0-100. If the score is available, record the score. If
only the risk level is available, record that.
Note 6: For Mammaprint, EndoPredict, and Breast Cancer Index, only record the risk level.
Code Description
00-99 Enter actual recurrence score
Note: Depending on the test, the range of values may be different
X1 Score 100
X2 Low risk
X3 Moderate [intermediate] risk
X4 High risk
X7 Test ordered, results not in chart
X8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit
error.)
X9 Not documented in medical record
Multigene Signature Results not assessed or unknown if assessed
Return to Schema ID Table

235 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
Oncotype Dx Tests
The recording of Oncotype Dx was previously collected in Multigene Signature Results and Multigene
Signature Method in CSv2. Oncotype Dx now has four data items.
3903: Oncotype Dx Recurrence Score-DCIS
3904: Oncotype Dx Recurrence Score-Invasive
3905: Oncotype Dx Risk Level-DCIS
3906: Oncotype Dx Risk Level-Invasive
Oncotype DX DCIS Score
Definition
The Oncotype DX DCIS score is a genomic test that estimates the likelihood of local recurrence (DCIS or
invasive) for a patient with DCIS. The results may be used clinically to evaluate benefits of radiation
therapy following surgery.
The Oncotype DX DCIS score, a numeric value from 0-100, is coded in NAACCR Data Item #3903.
Oncotype DX DCIS Risk Level, coded in NAACCR Data Item #3905, stratifies the Oncotype DX DCIS Score
into three risk levels
Low risk: Recurrence Score lower than 39: The DCIS has a lower risk of recurrence.
Intermediate Risk: Recurrence Score between 39 and 54: The DCIS has an intermediate risk of
recurrence.
High risk: Recurrence Score greater than 54: The DCIS has a higher risk of recurrence.
Additional Information
Source documents: Oncotype Dx DCIS laboratory report, other statements in medical record
For further information, see http://www.oncotypeiq.com/en-US/breast-cancer/patients-and-
caregivers/stage-0-dcis/about-the-test

236 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Oncotype DX Breast Recurrence Score
Definition
The Oncotype DX Breast Recurrence Score test (Oncotype DX) test is a genomic test that predicts the
risk of distant recurrence and likelihood of benefit chemotherapy for early stage breast cancers. It is
required for assigning prognostic stage in AJCC 8
th
edition for patients with T1-2 N0, M0, ER-positive,
HER2 negative breast cancers. Oncotype DX provides a quantitative score, based on a continuous scale
from 0-100, with higher scores reflecting higher risk of distant recurrence and higher likelihood of
chemotherapy benefit.
The numeric value of the recurrence score is coded in Data Item #3906. When the actual recurrence
score is not available, there is an option for coding recurrence scores stated as less than 11 or greater
than equal to 11 as this the cut point determined to be clinically relevant for stage group in AJCC8.
Oncotype DX Risk Level -Invasive, coded in NAACCR Data Item #3906, stratifies the Oncotype DX
recurrence score into three risk levels
Low risk: Recurrence Score result less than 18: The patient has a lower risk of having a
recurrence, assuming 5 years of hormonal therapy is given. Chemotherapy is likely to have little
or no benefit.
Intermediate Risk: Recurrence Score result between 18 and 30: The patient has a tumor that is
in the middle of the risk spectrum reflecting that biology is continuous and not all patients have
a low or a high recurrence risk, assuming 5 years of hormonal therapy is given. The likelihood of
distant recurrence and benefit from chemotherapy increases with an increase in the Recurrence
Score result.
High risk: Recurrence Score result greater than or equal to 31: The patient has a high risk of
distant recurrence, assuming 5 years of hormonal therapy and is likely to benefit from
chemotherapy.
Additional Information
Source documents: Oncotype Dx Breast Recurrence Score laboratory report, other statements
in medical record
For further information, see http://www.oncotypeiq.com/en-US/breast-cancer/healthcare-
professionals/oncotype-dx-breast-recurrence-score/about-the-test

237 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3903: Oncotype Dx Recurrence Score-DCIS
Item Length: 3
NAACCR Item #: 3903
XML Parent-NAACCR ID: Tumor-oncotypeDxRecurrenceScoreDcis
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Oncotype Dx Recurrence Score-DCIS is a numeric score of a genomic test to predict the likelihood of
distant recurrence of invasive breast cancer based on the assessment of 21 genes.
Rationale
Oncotype Dx Recurrence Score-DCIS is a Registry Data Collection Variable in AJCC. It is a new data item
for cases diagnosed 1/1/2018+.
See 3903-3906: Oncotype Dx Tests for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Oncotype Dx Recurrence Score-DCIS can be used to code this data item.
Note 2: The Oncotype Dx-DCIS recurrence score is reported as a whole number between 0 and 100.
Note 3: Record only the results of an Oncotype Dx-DCIS recurrence score in this data item. If some other
test is used for scoring, assign code XX9.
Note 4: In cases where Oncotype Dx-DCIS is reported on more than one in situ breast tumor specimen,
record the highest value.
Note 5: Code XX9 for LCIS tumors.
Note 6: If the only information available is the Oncotype Dx-DCIS Risk Level, assign XX7.
Note 7: Code this data item using the same report used to record 3905: Oncotype Dx Risk Level-DCIS.
Code Description
000-100 Enter actual recurrence score between 0 and 100
XX6 Not applicable: invasive case
XX7
Test
ordered
, results not in chart
XX8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XX8 will result in an edit error.)
XX9
Not documented in medical record
Oncotype Dx Recurrence Score-DCIS not assessed or unknown if assessed
Return to Schema ID Table

238 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3905: Oncotype Dx Risk Level-DCIS
Item Length: 1
NAACCR Item #: 3905
XML Parent-NAACCR ID: Tumor-oncotypeDxRiskLevelDcis
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Oncotype Dx Risk Level-DCIS stratifies Oncotype Dx recurrence scores into low, intermediate, and high
risk of local recurrence.
Rationale
Oncotype Dx Risk Level-DCIS is a Registry Data Collection Variable in AJCC. It is a new data item for cases
diagnosed 1/1/2018+.
See 3903-3906: Oncotype Dx Tests for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Oncotype Dx Risk Level-DCIS can be used to code this data item.
Note 2: The Oncotype Dx Risk Level-DCIS test stratifies scores into low, intermediate, and high risk of
distant recurrence. If only the score is stated, assign the risk level based on the score.
Note 3: Code 9 for LCIS tumors.
Note 4: Record only the results of an Oncotype Dx Risk Level-DCIS in this data item. If some other test is
used for scoring, assign code 9.
Note 5: Code this data item using the same report used to record 3903: Oncotype Dx Recurrence Score-
DCIS.
Code Description
0 Low risk (recurrence score 0-38)
1 Intermediate risk (recurrence score 39-54)
2 High risk (recurrence score greater than or equal to 55)
6 Not applicable: invasive case
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Oncotype Dx Risk Level-DCIS not assessed or unknown if assessed
Return to Schema ID Table

239 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3904: Oncotype Dx Recurrence Score-Invasive
Item Length: 3
NAACCR Item #: 3904
XML Parent-NAACCR ID: Tumor-oncotypeDxRecurrenceScoreInvasiv
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Oncotype Dx Recurrence Score-Invasive is a numeric score of a genomic test to predict the likelihood of
distant recurrence of invasive breast cancer based on the assessment of 21 genes.
Rationale
Oncotype Dx Recurrence Score-Invasive is a Registry Data Collection Variable in AJCC. It is a new data
item for cases diagnosed 1/1/2018+.
See 3903-3906: Oncotype Dx Tests for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Oncotype Dx Recurrence Score-Invasive score can be used to code this
data item.
Note 2: The Oncotype Dx-Invasive recurrence score is reported as a whole number between 0 and 100.
The actual recurrence score takes priority over codes XX4 and XX5.
Note 3: Record only the results of an Oncotype Dx-Invasive recurrence score in this data item. If some
other test is used for scoring, assign code XX9.
Note 4: Predicted Oncotype Dx Recurrence Score based on linear regression models and Magee
equations should not be reported in this field.
If the only information you have on Oncotype Dx is based on a linear regression model and
Magee score, code unknown
Code the results of a Magee score in the Multigene Data Items: Multigene Signature Method
[NAACCR Data Item #3894] and Multigene Signature Results [NAACCR Data Item #3895]
Note 5: In cases where Oncotype DX is reported on more than one breast tumor specimen, record the
highest value.
Note 6: Results from nodal or metastatic tissue may be used ONLY when there is no evidence of primary
tumor.
Note 7: Staging for Breast cancer now depends on the Oncotype-Dx-Invasive recurrence score. Score of
less than 11 indicates a pertinent cut off value for staging purposes.

240 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 8: If the only information available is the Oncotype Dx-Invasive Risk Level, assign XX7.
Note 9: Code this data item using the same report used to record 3906: Oncotype Dx Risk Level-Invasive.
Code Description
000-100 Enter actual recurrence score between 0 and 100
XX4 Stated as less than 11
XX5 Stated as equal to or greater than 11
XX6 Not applicable: in situ case
XX7
Test
ordered
, results not in chart
XX9
Not documented in medical
record
Oncotype Dx Recurrence Score-Invasive not assessed or unknown if assessed
Return to Schema ID Table

241 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3906: Oncotype Dx Risk Level-Invasive
Item Length: 1
NAACCR Item #: 3906
XML Parent-NAACCR ID: Tumor-oncotypeDxRiskLevelInvasive
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Oncotype Dx Risk Level-Invasive stratifies Oncotype Dx recurrence scores into low, intermediate, and
high risk of distant recurrence.
Rationale
Oncotype Dx Risk Level-Invasive is a Registry Data Collection Variable in AJCC. It is a new data item for
cases diagnosed 1/1/2018+.
See 3903-3906: Oncotype Dx Tests for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Oncotype Dx Risk Level-Invasive can be used to code this data item.
Note 2: The Oncotype Dx Risk Level-Invasive test stratifies scores into low, intermediate, and high risk of
distant recurrence. If only the score is stated, assign the risk level based on the score.
Note 3: Record only the results of an Oncotype Dx Risk Level-Invasive in this data item. If some other
test is used for scoring, assign code 9.
Note 4: Code this data item using the same report used to record 3904: Oncotype Dx Recurrence Score-
Invasive.
Code Description
0 Low risk (recurrence score 0-17)
1 Intermediate risk (recurrence score 18-30)
2 High risk (recurrence score greater than or equal to 31)
6 Not applicable: DCIS case
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Oncotype Dx Risk Level-Invasive not assessed or unknown if assessed
Return to Schema ID Table

242 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3863: Ki-67
Item Length: 5
NAACCR Item #: 3863
XML Parent-NAACCR ID: Tumor-ki67
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
Ki-67 (MIB-1) (Proliferative Index) is a marker of cell proliferation. A high value indicates a tumor that is
proliferating more rapidly. Codes and coding instructions for this data item are site-specific.
Rationale
Ki-67 (MIB-1) (Proliferative Index) is a Registry Data Collection Variable in AJCC. It was a new data item
for breast cases diagnosed 1/1/2018+. It will apply to neuroendocrine tumors (NET) of the
gastrointestinal tract (AJCC Chapters 29 – 34) for cases diagnosed 1/1/2021+. High Ki-67 is an adverse
prognostic factor and Ki-67 is a component of grade for these tumors. NCCN guidelines recommend
that tumor differentiation, mitotic rate and Ki-67 should be recorded in the pathology report for these
tumors.
Coding Instructions
Note 1: Physician statement of Ki-67 (MIB-1) can be used to code this data item.
Note 2: Ki-67 is a marker of cell proliferation. A high value indicates a tumor that is proliferating more
rapidly.
Note 3: Results from nodal or metastatic tissue may be used, ONLY when there is no evidence of
primary tumor.
Note 4: Ki-67 results are reported as the percentage cell nuclei that stain positive. As of early 2017
there are no established standards for interpretation of results or for cutoffs for positive and negative.
Examples:
Ki-67 reported as 14%. Code 14.0
Ki-67 reported as 8.6%. Code 8.6
Note 5: In cases where there are invasive and in situ components in the primary tumor and Ki-67 is done
on both, ignore the in situ results.
If Ki-67 is done on both the in situ and invasive components in the primary tumor, code the Ki-
67 value from the invasive component
If in situ and invasive components present and Ki-67 only done on the in situ component in the
primary tumor, code unknown

243 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0.0-100.0 0.0 to 100.0 percent positive: enter percent positive
XXX.7 Test done, actual percentage not stated
XXX.8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XXX.8 will result in an edit
error.)
XXX.9 Not documented in medical record
Ki-67 (MIB-1) not assessed or unknown if assessed
Return to Schema ID Table

244 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3882: LN Positive Axillary Level I-II
Item Length: 2
NAACCR Item #: 3882
XML Parent-NAACCR ID: Tumor-lnPositiveAxillaryLevel1To2
NAACCR Alternate Name: Lymph Nodes Positive Axillary Level I-II
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
This data item pertains to the number of positive ipsilateral level I and II axillary lymph nodes and
intramammary lymph nodes based on pathological information.
Rationale
Lymph Nodes Positive Axillary Level I-II can be collected by the surveillance community for breast
cancers. Prior to 2018, Breast SSF#3 was used for Lymph Nodes Positive Axillary Level I-II.
Definition
This data items records the low axillary (level I and intramammary) and mid-axillary (level II, also called
interpectoral or Rotter’s nodes).
This data item excludes level III (high axillary, also called apical or infraclavicular), internal mammary and
supraclavicular lymph nodes.
Do not confuse intramammary nodes, which are within breast tissue and are included in level I, with
internal mammary nodes, which are along the sternum.
This field is based on pathological examination of ipsilateral (same side as the primary cancer) level I and
II axillary lymph nodes, so pathological information is included even if the patient had neoadjuvant
therapy prior to lymph node removal.
Do not include lymph nodes containing only isolated tumor cells (ITCs—metastases less than 0.2 mm in
size) in the count of positive nodes.
Coding guidelines
Code 00 when all level I and II axillary lymph nodes are negative on pathological examination
Code the exact number of lymph nodes in the range 01 to 99 for the exact count of level I and II
axillary lymph nodes, or X1 if more than 99 level I and II axillary lymph nodes are positive
Code X5 if level I and II axillary lymph nodes were positive, but the number is not specified
Code X6 if there was only a positive aspiration of level I or II axillary lymph node(s)
Code X9 when
o No axillary nodes were examined
o Axillary dissection was performed but no axillary lymph nodes were found

245 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Clinical diagnosis only (no axillary lymph nodes were removed)
o Unknown whether axillary lymph nodes are positive
Additional information
Required for Staging: EOD only.
Source documents pathology report
Coding Instructions and Codes
Note 1: Physician statement of number of positive ipsilateral Level I-II axillary nodes can be used to code
this data item, when no other specific information is available.
Note 2: Include only the number of positive ipsilateral level I and II axillary lymph nodes and
intramammary lymph nodes in this field. Intramammary nodes, located within the breast, are not the
same as internal mammary nodes, located along the sternum.
Note 3: This field is based on microscopic information only. If no ipsilateral axillary nodes are examined,
or if an ipsilateral axillary lymph node drainage area is removed but no lymph nodes are found, code X9.
Note 4: For cases where neoadjuvant therapy is administered
If clinical nodal involvement is more extensive, include only those nodes that are positive during
clinical workup
o Positive nodes can be from an FNA, core biopsy or sentinel lymph node biopsy
Example: Patient with positive FNA of axillary lymph node, neoadjuvant therapy
administered. Lymph node dissection revealed negative lymph nodes. Code X6
for the positive FNA.
If the post-neoadjuvant nodal involvement is more extensive, include only those nodes positive
during surgery
o Positive nodes can be from an FNA, core biopsy, sentinel lymph node biopsy or lymph
node dissection
Example: Patient with large breast mass, lymph node negative on clinical exam.
Neoadjuvant therapy administered. Mastectomy and sentinel lymph node
biopsy done, 1 of 2 SLN’s positive. Code 01.
Note 5: Lymph nodes with only isolated tumor cells (ITCs) are not counted as positive lymph nodes.
Only lymph nodes with metastases greater than 0.2 mm (micrometastases or larger) should be counted
as positive. If the pathology report indicates that axillary nodes are positive, but size of the metastases is
not stated, assume the metastases are greater than 0.2 mm and code the lymph nodes as positive in this
field.
Note 6: When positive ipsilateral axillary lymph nodes are coded in this field, the number of positive
ipsilateral axillary lymph nodes must be less than or equal to the number coded in Regional Nodes
Positive (i.e., the number of positive ipsilateral axillary nodes will always be a subset of the number of
positive regional nodes.)

246 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
00 All ipsilateral axillary nodes examined negative
01
-
99
1
-
99 nodes positive
(Exact number of nodes positive)
X1 100 or more nodes positive
X5 Positive nodes, number unspecified
X6 Positive aspiration or needle core biopsy of lymph node(s)
X8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9
Not documented in medical record
Level I-II axillary nodes not assessed or unknown if assessed
Return to Schema ID Table

247 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00480: Breast (2018+)
3922: Response to Neoadjuvant Therapy
Item Length: 1
NAACCR Item #: 3922
XML Parent-NAACCR ID: Tumor-responseToNeoadjuvantTherapy
NAACCR Alternate Name: None
Active years: 2018+
Schemas(s):
00480: Breast (2018+)
Description
This data item records the physician’s statement of response to neoadjuvant chemotherapy.
Rationale
Response to Neoadjuvant Therapy is a Registry Data Collection Variable in AJCC. It was previously
collected as Breast, CS SSF #21.
Definition
Neoadjuvant therapy is defined as systemic or radiation treatment administered prior to surgery in an
attempt to shrink the tumor or destroy regional metastases. This data item documents whether that
neoadjuvant therapy was successful.
This data item is coded based on the clinician’s statement regarding response to neoadjuvant
therapy. Do not try to interpret or infer a response based on the medical record. As a guide for the
clinician, the definitions below are from the AJCC Cancer Staging Manual, 8
th
edition.
The registrar should not use these definitions to code this field
Complete Response (CR) – absence of invasive carcinoma in breast and lymph nodes; must be
determined by microscopic evaluation of tissues; residual in situ cancer at primary site
Partial Response (PR) – a decrease in T and/or N category compared to pretreatment value and
no increase, using same method of evaluation as baseline value; residual tumor in lymph nodes
of any size
No Response (NR) – no apparent change in the T or N category compared to pretreatment value,
or an increase in T or N value at time of y pathological examination
Coding guidelines
Code 0 if there is no neoadjuvant therapy given
o This includes in situ (behavior /2) cases
Code 1 for a Residual Cancer Burden (RCB) result of '0' or an RCB Class of pCR (pathological
complete response).
Code 9 when
o there is no statement of complete, partial or no response by the clinician or when the
response is not documented in the medical record

248 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
For further information, refer to the Breast or Breast Biomarker Reporting cancer protocols
published by the College of American Pathologists for the AJCC Staging System Breast
Other names: treatment effect
Coding Instructions and Codes
Note 1: Clinician statement of Response to Neoadjuvant Therapy (“treatment effect”) must be used to
code this data item.
Note 2: The clinician’s statement may be based on pathology reports, imaging, and other clinical
findings.
Note 3: Code 1 is to be used only when the physician states the response is “total” or “complete.”
Code Description
0
Neoadjuvant therapy not given
Non-invasive neoplasm (behavior /2)
1 Stated as complete response (CR)
2 Stated as partial response (PR)
3 Stated as response to treatment, but not noted if complete or partial
4 Stated as no response (NR)
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit
error.)
9 Not documented in medical record
Response to neoadjuvant therapy not assessed or unknown if assessed
Return to Schema ID Table
249 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
FEMALE REPRODUCTIVE ORGANS

250 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
3836: FIGO
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00500: Vulva (FIGO: Vulva) (2018+)
00510: Vagina (FIGO: Vagina) (2018+)
00520: Cervix Uteri (2018-2020) (FIGO: Cervix) (2018+)
09520: Cervix Uteri (2021+) (FIGO: Cervix) (2018+)
00528: Cervix Sarcoma (2021+) (FIGO Stage (Sarcoma)) (2018+)
00530: Carcinoma and Carcinosarcoma (FIGO: Corpus Carcinoma and Carcinosarcoma) (2018+)
00541: Corpus Sarcoma (FIGO Stage (Adenosarcoma) (2018+)
00542: Corpus Adenosarcoma (FIGO Stage (Sarcoma)) (2018+)
00551: Ovary (FIGO: Ovary, Fallopian Tube, and Peritoneal Carcinoma) (2018+)
00552: Primary Peritoneal Carcinoma (FIGO: Ovary, Fallopian Tube, and Peritoneal Carcinoma)
(2018+)
00553: Fallopian Tube (FIGO: Ovary, Fallopian Tube, and Peritoneal Carcinoma) (2018+)
00560: Placenta (FIGO: Gestational Trophoblastic Tumors (Placenta)) (2018+)
Description
Federation Internationale de Gynecologie et d'Obstetrique (FIGO) is a staging system for female
reproductive cancers.
Rationale
FIGO stage is a Registry Data Collection Variable in AJCC for the female genital cancers. This data item
was previously collected for the female genital cancers as: Vulva SSF #10, Vagina SSF #1, Cervix SSF #1,
Corpus Carcinoma SSF #1, Corpus Sarcoma SSF #1, Ovary SSF #2, Fallopian Tube SSF #1, Peritoneum
Female Genital SSF #1, and Placenta SSF #2.
Definition
FIGO is the French acronym for the Federation Internationale de Gynecologie et d’Obstetrique, the
worldwide organization of obstetricians and gynecologists who maintain the international staging
systems for female genital organs. In English, the organization is the International Federation of
Gynecology and Obstetrics. The FIGO staging system has been adapted into the AJCC staging
manual. FIGO uses Roman numerals and subscripts to define a stage. There is no T, N, or M descriptor
with FIGO stage, only a stage group. For example, FIGO Stage IA is equivalent to T1a, FIGO Stage III can
be either T3_ or N1, and FIGO Stage IV is M1.
Definitions of the various FIGO stages vary from primary to primary, but the structure is similar
throughout. FIGO no longer includes an in situ stage (Tis, Stage 0). For in situ tumors, code the following
Code 97: Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
Note: Do not confuse FIGO stage with FIGO grade.

251 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Structure of Codes
For all sites, the structure of the FIGO data items is the same, although not every system uses every
possible FIGO code and the actual codes used are not the same for all systems.
Coding guidelines
Code the FIGO stage as stated in the medical record. When lymph node(s) is/are clinically or
pathologically positive or metastasis is present, make sure that the FIGO stage reflects the combination
of T, N, and M and NOT just the T. If a stage group is stated but it does not specify that it is a FIGO stage,
assume that it is a FIGO stage and code it. Do not attempt to code FIGO stage based only on T, N, and M.
If you cannot make a determination of stage based on the previous information, code 99.
1 FIGO Stage I (all systems)
1B2 FIGO Stage IB2 (cervix only)
2 FIGO Stage II (all systems)
3A12 FIGO Stage IIIA1ii (ovary, fallopian tube, and primary peritoneal carcinoma only)
4 FIGO Stage IV (all systems)
99: FIGO Stage unknown, FIGO stage not assessed or unknown if FIGO stage assessed
Additional Information
Source documents: clinician’s notes, consultant notes, pathology report, radiation therapy
notes
FIGO Stage: Summary of Schemas
Code Description Vulva
Vagina Cervix Corpus
Sarcoma
Corpus
Adeno-
Sarcoma
Corpus
Carcinoma
Ovary,
FT, PPC
Placenta
1 FIGO Stage I X X X X X X X* X
1A FIGO Stage IA X X X X X X*
1A1 FIGO Stage IA1 X
1A2 FIGO Stage IA2 X
1B FIGO Stage IB X X X X X X*
1B1 FIGO Stage IB1 X
1B2 FIGO Stage IB2 X
1B3 FIGO Stage IB3 X**
1C FIGO Stage IC X X*
1C1 FIGO Stage IC1 X*
1C2 FIGO Stage IC2 X*
1C3
FIGO Stage IC3
X
*
2
FIGO Stage II
X
X
X
X
X
X
X
X
2A FIGO Stage IIA X X X X
2A1 FIGO Stage IIA1 X
2A2 FIGO Stage IIA2 X
2B FIGO Stage IIB X X X
3 FIGO Stage III X X X X X X X X
3A FIGO Stage IIIA X X X X X X
3A1 FIGO Stage IIIA1 X

252 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description Vulva
Vagina Cervix Corpus
Sarcoma
Corpus
Adeno-
Sarcoma
Corpus
Carcinoma
Ovary,
FT, PPC
Placenta
3A11 FIGO Stage IIIA1i X
3A12 FIGO Stage IIIA1ii X
3A2 FIGO Stage IIIA2 X
3B FIGO Stage IIIB X X X X X X
3C FIGO Stage IIIC X X X X X X
3C1 FIGO Stage IIIC1 X** X
3C2 FIGO Stage IIIC2 X** X
4 FIGO Stage IV X X X X X X X X
4
A
FIGO Stage IVA
X
X
X
X
X
X
X
4B FIGO Stage IVB X X X X X X X
*Not applicable for Primary Peritoneal Carcinoma
** Cervix Version 9 only (effective 1/1/2021 forward)
In addition to the codes listed above, the following codes are also applicable
Code
Description
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)*
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
*Not applicable for the Cervix Sarcoma, Corpus Adenosarcoma, and Corpus Sarcoma
Return to Schema ID Table

253 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN
Sites
Definition
In addition to assigning the N categories for cervix, vagina and vulva cancers, the collection of specific
lymph nodes and how they were assessed is important.
Status refers to positive or negative involvement
Assessment is the method by which the nodal status was determined
There are 6 data items that collect information on regional lymph nodes. Three data items collect the
individual status (positive, negative, unknown) involvement of femoral-inguinal, para-aortic and pelvic
lymph nodes. There are 3 assessment data items that collect individual status information on the 3
regional lymph node groups.
3871: LN Assessment Method Femoral-Inguinal
3872: LN Assessment Method Para-Aortic
3873: LN Assessment Method Pelvic
3957: LN Status: Pelvic
3958: LN Status: Para-Aortic
3959: LN Status: Femoral-Inguinal
There are 2 data items that collect information on distant lymph nodes. One data item collects the
status (positive, negative, unknown) involvement of mediastinal and scalene distant lymph nodes. The
other data item collects the assessment method.
3874: LN Distant Assessment Method
3875: LN Distant: Mediastinal, Scalene
For the 4 status fields, the data items have a basic set up
Code 0 when all lymph nodes are negative
Multiple codes are available to record single or multiple involvement of lymph nodes
Code 9 when
o Not documented in medical record
o Regional/Distant lymph nodes not evaluated (assessed)
o Unknown if regional/distant lymph nodes evaluated (assessed)
For the 4 methods fields, the codes are the same
Code 0 when there is physical exam or imaging only
Code 1 when there is an incisional biopsy or FNA
Code 2 when there is an excisional biopsy or lymph node resection
Code 7 when lymph nodes are assessed, but it is unknown how
Code 9 when
o Not documented in medical record
o Regional/Distant lymph nodes not evaluated (assessed)
o Unknown if regional/distant lymph nodes evaluated (assessed)

254 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vulva (2018+)
3836: FIGO: Vulva
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Note 4: The FIGO stage definitions do not include Stage 0 (Tis).
Code 97 for any non-invasive neoplasm (behavior /2)
Code Description
1 FIGO Stage I
1A FIGO Stage IA
1B FIGO Stage IB
2 FIGO Stage II
3
FIGO Stage III
3A
FIGO Stage IIIA
3B FIGO Stage IIIB
3C FIGO Stage IIIC
4 FIGO Stage IV
4A FIGO Stage IVA
4B FIGO Stage IVB
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
98
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99
Not documented in medical
record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

255 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vulva (2018+)
3959: LN Status: Femoral-Inguinal
Item Length: 1
NAACCR Item #: 3959
XML Parent-NAACCR ID: Tumor-lnStatusFemoralInguinal
NAACCR Alternate Name: Lymph Nodes Status: Femoral-Inguinal
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
Description
This data item describes the status of femoral-inguinal lymph nodes associated with certain female
genital cancers.
Rationale
Specific regional lymph node involvement is listed as a Registry Data Collection Variable in AJCC. This
information was previously collected as Vulva, SSF #14
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of the femoral-inguinal status can be used to code this data item when no
other information is available.
Note 2: The following are femoral-inguinal nodes
Femoral
Inguinal, NOS
o Inguinofemoral (groin)
o Node of Cloquet or Rosenmuller (highest deep inguinal)
o Superficial inguinal
Note 3: If there is no mention of femoral-inguinal lymph node involvement in the area being imaged,
biopsied, or in the surgical field and there is no mention of involvement, then assume that the femoral-
inguinal lymph nodes are negative.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Code 9 when there is no imaging, biopsy, surgical workup, or a physical exam only.
Note 6: The assessment method is recorded in 3871: LN Assessment Method Femoral-Inguinal.

256 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Negative femoral-inguinal lymph nodes
Non-invasive neoplasm (behavior /2)
1 Positive femoral-inguinal lymph nodes
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Femoral-Inguinal lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

257 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vulva (2018+)
3871: LN Assessment Method Femoral-Inguinal
Item Length: 1
NAACCR Item #: 3871
XML Parent-NAACCR ID: Tumor-lnAssessMethodFemoralInguinal
NAACCR Alternate Name: Lymph Nodes Assessment Method Femoral-Inguinal
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
0510: Vagina (2018+)
Description
This data item describes the method used to assess involvement of femoral-inguinal lymph nodes
associated with certain female genital cancers.
Rationale
Method of assessment of regional nodal status is listed as a Registry Data Collection Variable in the AJCC
GYN chapters. This data item was previously collected as Vulva, SSF #15.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of femoral-inguinal assessment method can be used to code this data item
when no other information is available.
Note 2: The following are femoral-inguinal nodes
Femoral
Inguinal, NOS
o Inguinofemoral (groin)
o Node of Cloquet or Rosenmuller (highest deep inguinal)
o Superficial inguinal
Note 3: Assign the highest applicable code (0-2) in the case of multiple assessments.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Code 0 when there is only imaging or a physical exam.
Note 6: The status results are recorded in 3959: LN Status: Femoral-Inguinal.

258 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Radiography, imaging
(Ultrasound (US), computed tomography scan (CT), magnetic resonance imaging (MRI),
positron emission tomography scan (PET))
Physical exam only
1 Incisional biopsy; fine needle aspiration (FNA)
2 Lymphadenectomy
Sentinel node biopsy
Excisional biopsy or resection with microscopic confirmation
7 Femoral-inguinal lymph node(s) assessed, unknown assessment method
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not
documented in medical record
Femoral-inguinal lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

259 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vulva (2018+)
3957: LN Status: Pelvic
Item Length: 1
NAACCR Item #: 3957
XML Parent-NAACCR ID: Tumor-lnStatusPelvic
NAACCR Alternate Name: Lymph Node Status: Pelvic
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the status of pelvic lymph nodes associated with certain female genital cancers.
Rationale
Specific regional lymph node involvement is listed as a Registry Data Collection Variable in AJCC. This
information was previously collected as Vulva, SSF #12
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding instructions and Codes
Note 1: Physician statement of pelvic status can be used to code this data item when no other
information is available.
Note 2: For Vulva, pelvic lymph nodes are distant.
Note 3: The following are pelvic nodes
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)
Paracervical
Parametrial
Pelvic, NOS
Sacral, NOS
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Uterosacral
Note 4: If there is no mention of pelvic lymph node involvement in the area being imaged, biopsied, or
in the surgical field and there is no mention of involvement, then assume that the pelvic lymph nodes
are negative.
Note 5: For this data item, do not include isolated tumor cells (ITCs).

260 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 6: Code 9 when there is no imaging, biopsy, surgical workup, or a physical exam only.
Note 7: The assessment method is recorded in 3873: LN Assessment Method Pelvic.
Code Description
0 Negative pelvic lymph nodes
Non-invasive neoplasm (behavior /2)
1 Positive pelvic lymph nodes
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter,
use of code 8 may result in an edit error.)
9 Not documented in medical record
Pelvic lymph node(s) not assessed or unknown if
assessed
Return to Schema ID Table

261 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vulva (2018+)
3873: LN Assessment Method Pelvic
NAACCR Item #: 3873
XML Parent-NAACCR ID: Tumor-lnAssessMethodPelvic
NAACCR Alternate Name: Lymph Nodes Assessment Method Pelvic
Active years: 2018+
Schemas(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the method used to assess involvement of pelvic lymph nodes associated with
certain female genital cancers.
Rationale
Method of assessment of regional nodal status is listed as a Registry Data Collection Variable in the AJCC
GYN chapters. This data item was previously collected as Vulva, SSF #13.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of pelvic assessment method can be used to code this data item when no
other information is available.
Note 2: For Vulva, pelvic lymph nodes are distant.
Note 3: The following are pelvic nodes
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)
Paracervical
Parametrial
Pelvic, NOS
Sacral, NOS
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Uterosacral
Note 4: Assign the highest applicable code (0-2) in the case of multiple assessments.
Note 5: For this data item, do not include isolated tumor cells (ITCs).
Note 6: Code 0 when there is only imaging or a physical exam.

262 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 7: The assessment results are recorded in 3957: LN Status: Pelvic.
Code Description
0
Radiography, imaging
(Ultrasound (US), computed tomography scan (CT), magnetic resonance imaging (MRI),
positron emission tomography scan (PET))
Physical exam only
1 Incisional biopsy; fine needle aspiration (FNA)
2 Lymphadenectomy
Sentinel node biopsy
Excisional biopsy or resection with microscopic confirmation
7 Pelvic lymph node(s) assessed, unknown assessment method
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Pelvic lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

263 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vulva (2018+)
3881: LN Laterality
Item Length: 1
NAACCR Item #: 3881
XML Parent-NAACCR ID: Tumor-lnLaterality
NAACCR Alternate Name: Lymph Nodes Laterality
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
Description
This data item describes whether positive regional lymph nodes are unilateral or bilateral.
Rationale
Laterality of regional node metastasis is a Registry Data Collection Variable in AJCC. This data item was
previously collected as Vulva, CS SSF #11.
Definition
This data item records the appropriate description of involved regional lymph nodes, specifically
whether they are unilateral or bilateral involvement.
Coding guidelines
Code the appropriate description of involved regional lymph nodes
Code 0 when all regional lymph nodes are negative
Code 1 when
o all positive regional nodes are ipsilateral
o involved lymph nodes are described as unilateral
Code 2 when
o at least one regional lymph node is involved on each side of the pelvis
o involvement is described as bilateral or contralateral
Code 3 when regional lymph node(s) are described as positive but the laterality of the involved
nodes is unknown
Code 9 when
o Lymph nodes were not examined or assessed
o there is no information in the medical record about regional lymph node involvement
o the status of regional lymph nodes is unknown
Additional Information
Source documents: pathology report, imaging, physical exam, other statement in record

264 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note: Physician statement of lymph node laterality can be used to code this data item when no other
information is available.
Code Description
0 No regional lymph node involvement
Non-invasive neoplasm (behavior /2)
1 Unilateral - all positive regional nodes with same laterality
OR only one regional node positive
2 Bilateral - positive bilateral regional lymph nodes
3 Laterality unknown - positive regional lymph nodes with unknown laterality
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Lymph node laterality not assessed or unknown if assessed
Return to Schema ID Table

265 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina (2018+)
3836: FIGO: Vagina
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00510: Vagina (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Note 4: The FIGO stage definitions do not include Stage 0 (Tis).
Code 97 for any non-invasive neoplasm (behavior /2)
Code Description
1 FIGO Stage I
2 FIGO Stage II
3 FIGO Stage III
4 FIGO Stage IV
4A
FIGO Stage IVA
4B
FIGO Stage IVB
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

266 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina (2018+)
3959: LN Status: Femoral-Inguinal
Item Length: 1
NAACCR Item #: 3959
XML Parent-NAACCR ID: Tumor-lnStatusFemoralInguinal
NAACCR Alternate Name: Lymph Nodes Status: Femoral-Inguinal
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
Description
This data item describes the status of femoral-inguinal lymph nodes associated with certain female
genital cancers.
Rationale
Specific regional lymph node involvement is listed as a Registry Data Collection Variable in AJCC. This
information was previously collected as Vulva, SSF #14
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of the femoral-inguinal status can be used to code this data item when no
other information is available.
Note 2: Code this data item for the lower third of the vagina only.
Code 9 for upper two thirds of the vagina, or unknown whether it’s the lower third or upper two
thirds
Note 3: The following are femoral-inguinal nodes
Femoral
Inguinal, NOS
o Inguinofemoral (groin)
o Node of Cloquet or Rosenmuller (highest deep inguinal)
o Superficial inguinal
Note 4: If there is no mention of femoral-inguinal lymph node involvement in the area being imaged,
biopsied, or in the surgical field and there is no mention of involvement, then assume that the femoral-
inguinal lymph nodes are negative.
Note 5: For this data item, do not include isolated tumor cells (ITCs).
Note 6: Code 9 when there is no imaging, biopsy, surgical workup, or a physical exam only.
Note 7: The assessment method is recorded in 3871: LN Assessment Method Femoral-Inguinal.

267 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Negative femoral-inguinal lymph nodes
Non-invasive neoplasm (behavior /2)
1 Positive femoral-inguinal lymph nodes
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Femoral-Inguinal lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

268 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00500: Vagina (2018+)
3871: LN Assessment Method Femoral-Inguinal
Item Length: 1
NAACCR Item #: 3871
XML Parent-NAACCR ID: Tumor-lnAssessMethodFemoralInguinal
NAACCR Alternate Name: Lymph Nodes Assessment Method Femoral-Inguinal
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
Description
This data item describes the method used to assess involvement of femoral-inguinal lymph nodes
associated with certain female genital cancers.
Rationale
Method of assessment of regional nodal status is listed as a Registry Data Collection Variable in the AJCC
GYN chapters. This data item was previously collected as Vulva, SSF #15.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of femoral-inguinal assessment method can be used to code this data item
when no other information is available.
Note 2: The following are femoral-inguinal nodes
Femoral
Inguinal, NOS
o Inguinofemoral (groin)
o Node of Cloquet or Rosenmuller (highest deep inguinal)
o Superficial inguinal
Note 3: Code this data item for the lower third of the vagina only.
Code 9 for upper two thirds of the vagina, or unknown whether it’s the lower third or upper two
thirds
Note 4: Assign the highest applicable code (0-2) in the case of multiple assessments.
Note 5: For this data item, do not include isolated tumor cells (ITCs).
Note 6: Code 0 when there is only imaging or a physical exam.
Note 7: The status results are recorded in 3959: LN Status: Femoral-Inguinal.

269 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Radiography, imaging
(Ultrasound (US), computed tomography scan (CT), magnetic resonance imaging (MRI),
positron emission tomography scan (PET))
Physical exam only
1 Incisional biopsy; fine needle aspiration (FNA)
2 Lymphadenectomy
Sentinel node biopsy
Excisional biopsy or resection with microscopic confirmation
7 Femoral-inguinal lymph node(s) assessed, unknown assessment method
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Femoral-inguinal lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

270 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina (2018+)
3958: LN Status: Para-Aortic
Item Length: 1
NAACCR Item #: 3958
XML Parent-NAACCR ID: Tumor-lnStatusParaAortic
NAACCR Alternate Name: Lymph Node Status: Para-aortic
Active years: 2018+
Schema(s):
00510: Vagina (2018+)
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the status of para-aortic lymph nodes associated with certain female genital
cancers.
Rationale
Specific regional lymph node involvement is listed as a Registry Data Collection Variable in AJCC. This
information was previously collected as Vagina SSF #4
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of para-aortic status can be used to code this data item when no other
information is available.
Note 2: The following are para-aortic nodes
Aortic
Lateral aortic/lumbar aortic
Para-aortic, NOS
Periaortic
Note 3: If there is no mention of para-aortic lymph node involvement in the area being imaged,
biopsied, or in the surgical field and there is no mention of involvement, then assume that the para-
aortic lymph nodes are negative.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Code 9 when there is no imaging, biopsy, surgical workup, or a physical exam only.
Note 6: The assessment method is recorded in 3872: LN Assessment Method Para-Aortic.

271 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code
Description
0 Negative para-aortic lymph nodes
Non-invasive neoplasm (behavior /2)
1 Positive para-aortic lymph nodes
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter,
use of code 8 may result in an edit error.)
9 Not documented in medical record
Para-aortic lymph node(s) not assessed or unknown if
assessed
Return to Schema ID Table

272 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina (2018+)
3872: LN Assessment Method Para-Aortic
Item Length: 1
NAACCR Item #: 3872
XML Parent-NAACCR ID: Tumor-lnAssessMethodParaaortic
NAACCR Alternate Name: Lymph Nodes Assessment Method Para-aortic
Active years: 2018+
Schema(s):
00510: Vagina (2018+)
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the method used to assess involvement of para-aortic lymph nodes associated
with certain female genital cancers.
Rationale
Method of assessment of regional nodal status is listed as a Registry Data Collection Variable in the AJCC
GYN chapters. This data item was previously collected as Vagina, CS SSF #5.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of para-aortic assessment of nodal status for para-aortic nodes can be used
to code this data item when no other information is available.
Note 2: The following are para-aortic nodes
Aortic
Lateral aortic/lumbar aortic
Para-aortic, NOS
Periaortic
Note 3: Assign the highest applicable code (0-2) in the case of multiple assessments.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Code 0 when there is only imaging or a physical exam.
Note 6: The assessment results are recorded in 3958: LN Status: Para-Aortic.
Code Description
0
Radiography, imaging
(Ultrasound (US), computed tomography scan (CT), magnetic resonance imaging (MRI),
positron emission tomography scan (PET))
Physical exam only

273 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
1 Incisional biopsy; fine needle aspiration (FNA)
2 Lymphadenectomy
Sentinel node biopsy
Excisional biopsy or resection with microscopic confirmation
7 Para-aortic lymph node(s) assessed, unknown assessment method
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Para-aortic lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

274 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina
3957: LN Status: Pelvic
Item Length: 1
NAACCR Item #: 3957
XML Parent-NAACCR ID: Tumor-lnStatusPelvic
NAACCR Alternate Name: Lymph Node Status: Pelvic
Active years: 2018+
Schema(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the status of pelvic lymph nodes associated with certain female genital cancers.
Rationale
Specific regional lymph node involvement is listed as a Registry Data Collection Variable in AJCC. This
information was previously collected as Vulva, SSF #12
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding instructions and Codes
Note 1: Physician statement of pelvic status can be used to code this data item when no other
information is available.
Note 2: The following are pelvic nodes
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)
Paracervical
Parametrial
Pelvic, NOS
Sacral, NOS
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Uterosacral
Note 3: If there is no mention of pelvic lymph node involvement in the area being imaged, biopsied, or
in the surgical field and there is no mention of involvement, then assume that the pelvic lymph nodes
are negative.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Code 9 when there is no imaging, biopsy, surgical workup, or a physical exam only.

275 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 6: The assessment method is recorded in 3873: LN Assessment Method Pelvic.
Code Description
0 Negative pelvic lymph nodes
Non-invasive neoplasm (behavior /2)
1 Positive pelvic lymph nodes
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter,
use of code 8 may result in an edit error.)
9 Not documented in medical record
Pelvic lymph node(s) not assessed or unknown if
assessed
Return to Schema ID Table

276 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina (2018+)
3873: LN Assessment Method Pelvic
NAACCR Item #: 3873
XML Parent-NAACCR ID: Tumor-lnAssessMethodPelvic
NAACCR Alternate Name: Lymph Nodes Assessment Method Pelvic
Active years: 2018+
Schemas(s):
00500: Vulva (2018+)
00510: Vagina (2018+)
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the method used to assess involvement of pelvic lymph nodes associated with
certain female genital cancers.
Rationale
Method of assessment of regional nodal status is listed as a Registry Data Collection Variable in the AJCC
GYN chapters. This data item was previously collected as Vulva, SSF #13.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of pelvic assessment method can be used to code this data item when no
other information is available.
Note 2: The following are pelvic nodes
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)
Paracervical
Parametrial
Pelvic, NOS
Sacral, NOS
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Uterosacral
Note 3: Assign the highest applicable code (0-2) in the case of multiple assessments.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Code 0 when there is only imaging or a physical exam.
Note 6: The assessment results are recorded in 3957: LN Status: Pelvic.

277 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Radiography, imaging
(Ultrasound (US), computed tomography scan (CT), magnetic resonance imaging (MRI),
positron emission tomography scan (PET))
Physical exam only
1 Incisional biopsy; fine needle aspiration (FNA)
2 Lymphadenectomy
Sentinel node biopsy
Excisional biopsy or resection with microscopic confirmation
7 Pelvic lymph node(s) assessed, unknown assessment method
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Pelvic lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

278 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina (2018+)
3875: LN Distant: Mediastinal, Scalene
Item Length: 1
NAACCR Item #: 3875
XML Parent-NAACCR ID: Tumor-lnDistantMediastinalScalene
NAACCR Alternate Name: Lymph Nodes Distant: Mediastinal, Scalene
Active years: 2018+
Schema(s):
00510: Vagina
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the status of Distant (mediastinal, scalene) nodes associated with certain
female genital cancers.
Rationale
Specific distant lymph node involvement is listed as a Registry Data Collection Variable in the AJCC. This
data was previously collected as Vagina, CS SSF #6.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of mediastinal and scalene nodal status can be used to code this data item
when no other information is available.
Note 2: Assign the highest applicable code (1-3) in the case of positive nodes.
Note 3: If a nodal station is in the area being imaged, biopsied, or in the surgical field and there is no
mention of involvement, then assume that specific nodal station is negative.
Note 4: Code 9 when there is no imaging, biopsy, surgical workup, or a physical exam only.
Note 5: The assessment method is recorded in 3874: LN Distant Assessment Method.
Code Description
0 Negative mediastinal and scalene lymph nodes
Non-invasive neoplasm (behavior /2)
1 Positive mediastinal lymph nodes
2 Positive scalene lymph nodes
3 Positive mediastinal and scalene lymph nodes
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Mediastinal and scalene lymph nodes not assessed or unknown if assessed
Return to Schema ID Table

279 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00510: Vagina
3874: LN Distant Assessment Method
Item Length: 1
NAACCR Item #: 3874
XML Parent-NAACCR ID: Tumor-lnDistantAssessMethod
NAACCR Alternate Name: Lymph Nodes Distant Assessment Method
Active years: 2018+
Schema(s):
00510: Vagina
00520: Cervix (2018-2020)
09520: Cervix (2021+)
Description
This data item describes the method used to assess involvement of Distant (mediastinal, scalene) nodes
associated with certain female genital cancers.
Rationale
Method of assessment of distant nodal status is listed as a Registry Data Collection Variable in the AJCC
GYN chapters. This data item was previously collected as Vagina, CS SSF #7.
See Lymph Node Assessment Methods and Status for Regional and Distant Lymph Nodes in GYN Sites
for additional information
Coding Instructions and Codes
Note 1: Physician statement of Mediastinal and Scalene assessment method can be used to code this
data item when no other information is available.
Note 2: Assign the highest applicable code (0-2) in the case of multiple assessments.
Note 3: Code 0 when there is only imaging or a physical exam.
Note 4: The assessment results are recorded in 3875: LN Distant: Mediastinal, Scalene.
Code Description
0 Radiography, imaging
(Ultrasound (US), computed tomography scan (CT), magnetic resonance imaging (MRI),
positron emission tomography scan (PET))
Physical exam only
1 Incisional biopsy; fine needle aspiration (FNA)
2 Lymphadenectomy
Excisional biopsy or resection with microscopic confirmation
7 Distant lymph node(s) assessed, unknown assessment method
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Distant lymph node(s) not assessed or unknown if assessed
Return to Schema ID Table

280 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00520: Cervix (2018-2020)
See 00510: Vagina (2018+)
3958: LN Status: Para-Aortic
3872: LN Assessment Method Para-Aortic
3957: LN Status: Pelvic
3873: LN Assessment Method Pelvic
3874: LN Distant Assessment Method
3875: LN Distant: Mediastinal, Scalene
Return to Schema ID Table

281 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00520: Cervix (2018-2020)
3836: FIGO: Cervix
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00520: Cervix (2018-2020)
009520: Cervix (2021+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Note 4: The FIGO stage definitions do not include Stage 0 (Tis).
Code 97 for any non-invasive neoplasm (behavior /2)
Code Description
1 FIGO Stage I
1A FIGO Stage IA
1A1
FIGO Stage
IA1
1A2
FIGO Stage IA2
1B FIGO Stage IB
1B1 FIGO Stage IB1
1B2 FIGO Stage IB2
1B3 FIGO Stage IB3
2 FIGO Stage II
2A FIGO Stage IIA
2A1 FIGO Stage IIA1
2A2 FIGO Stage IIA2
2B FIGO Stage IIB
3 FIGO Stage III
3A FIGO Stage IIIA
3B FIGO Stage IIIB
3C FIGO Stage IIIC
3C1 FIGO Stage IIIC1
3C2
FIGO Stage IIIC2
4
FIGO Stage IV

282 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
4A FIGO Stage IVA
4B FIGO Stage IVB
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

283 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
09520: Cervix (2021+)
See 00510: Vagina (2018+)
3958: LN Status: Para-Aortic
3872: LN Assessment Method Para-Aortic
3957: LN Status: Pelvic
3873: LN Assessment Method Pelvic
3874: LN Distant Assessment Method
3875: LN Distant: Mediastinal, Scalene
See 00520: Cervix (2018-2020)
3836: FIGO: Cervix
Return to Schema ID Table

284 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
09520: Cervix (2021+)
3956: p16
Item Length: 1
NAACCR Item #: 3956
XML Parent-NAACCR ID: Tumor-p16
NAACCR Alternate Name: None
Active years: 2021+
Schema(s):
09520: Cervix (2021+)
09210: Anus (2023+)
Description
The p16 biomarker is overexpressed (produced) in response to HPV. It is therefore a surrogate marker
for HPV disease.
Rationale
Patients with HPV have a different survival or outcome so it is important to be able to distinguish this by
documenting the p16 results. Testing is performed by immunohistochemistry (IHC) which is inexpensive
and has near universal availability. It has an easily standardized interpretation. HPV testing is usually
performed through DNA testing which is more expensive and less widely available. HPV testing also has
technically more variability with the interpretation.
Definition
p16 is a tumor suppressor protein also known as cyclin-dependent kinase inhibitor 2A. The p16
biomarker is overexpressed (produced) in response to HPV. It is therefore a surrogate marker for HPV
disease.
Coding Instructions and Codes
Note 1: This SSDI is effective for diagnosis years 2021+.
For cases diagnosed 2018-2020, leave this SSDI blank
Note 2: Code 0 for p16 expression of weak intensity or limited distribution.
Note 3: This data item must be based on testing results for p16 overexpression.
A statement of a patient being HPV positive or negative is not enough to code this data item
Testing for HPV by DNA, mRNA, antibody, or other methods should not be coded in this data
item
Do not confuse p16 with HPV 16, which is a specific strain of virus
Code Description
0 p16 Negative; Nonreactive
1 p16 Positive; Diffuse, Strong reactivity
8 Not applicable: Information not collected for this case
(If this time is required by your standard setter, use of code 8 will result in an edit
error)
9 Not tested for p16; Unknown
<Blank> N/A-Diagnosis year prior to 2021
Return to Schema ID Table

285 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00528: Cervix Sarcoma
See 00541: Corpus Sarcoma (2018+)
3836: FIGO Stage (Sarcoma)
See 00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3901: Number of Positive Para-Aortic Nodes
3899: Number of Examined Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3900: Number of Examined Pelvic Nodes
3911: Peritoneal Cytology
Return to Schema ID Table

286 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3836: FIGO: Corpus Carcinoma and Carcinosarcoma
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Note 4: For Endometrial intraepithelial carcinoma (EIC) (8380/2) and Serous endometrial intraepithelial
carcinoma (SEIC) (8441/2), assign the FIGO staged based on the managing physician's documentation of
FIGO. (See Note 1).
If FIGO stage for EIC or SEIC is not documented by the managing physician, code unknown (code
99)
Do not code 97 (in situ) for EIC or SEIC since FIGO does not have a Stage 0
If diagnosis is Endometrial intraepithelial neoplasia (EIN) (8380/2), code 97.
Note 5: Code 97 for any remaining in situ histologies (/2) since the FIGO stage definitions do not include
Stage 0.
Code Description
1 FIGO Stage I
1A FIGO Stage IA
1B FIGO Stage IB
2 FIGO Stage II
3 FIGO Stage III
3A
FIGO Stage IIIA
3B
FIGO Stage IIIB
3C FIGO Stage IIIC
3C1 FIGO Stage IIIC1
3C2 FIGO Stage IIIC2
4 FIGO Stage IV
4A FIGO Stage IVA
4B FIGO Stage IVB

287 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
98
Not
applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99
Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

288 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3899-3902: Number of Positive and Examined Para-Aortic and Pelvic Nodes
Definition
Involvement of regional and distant lymph nodes is an important prognostic factor for cancers of the
gynecologic organs. The following list shows the regional and common distant lymph nodes for GYN
cancers.
Para-aortic nodes
Aortic
Lateral aortic/lumbar aortic
Para-aortic, NOS
Periaortic
Pelvic nodes
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)
Paracervical
Parametrial
Pelvis, NOS
Sacral
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Presacral
o Uterosacral
For the Cervix sarcoma cases, there are 4 data items that record information on the number of
positive and examined para-aortic and pelvic lymph nodes. These data items should be coded from
the same procedure
3899: Number of Examined Para-Aortic Nodes
3900: Number of Examined Pelvic Nodes
3901: Number of Positive Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
Number of nodes positive must ALWAYS be less than or equal to number of nodes examined.
Additional Information
Source documents: pathology report
Return to Schema ID Table

289 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3901: Number of Positive Para-Aortic Nodes
Item Length: 2
NAACCR Item #: 3901
XML Parent-NAACCR ID: Tumor-numberOfPositiveParaAorticNodes
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00528: Cervical Sarcoma (2021+)
00530: Corpus Carcinoma and Carcinosarcoma
00541: Corpus Sarcoma
00542: Corpus Adenosarcoma
Description
Number of Positive Para-Aortic Nodes is the number of positive nodes based on para-aortic nodal
dissection.
Rationale
Number of Positive Para-Aortic Nodes is listed as a Registry Data Collection Variable in AJCC. This data
item was previously collected as Corpus, CS SSF #5.
Coding guidelines
Code 00 for when there are no positive nodes
Code the exact number of positive nodes 01-99
Code X1 for 100 or more positive nodes
Code X2 for positive nodes, but unknown how many
Code X6 for aspiration or core biopsy of para-aortic node(s) only
Code X9 when
o Not documented in the medical record
o Para-Aortic lymph nodes not evaluated (assessed)
o No lymph node dissection performed
o Unknown if Para-Aortic lymph nodes evaluated (assessed)
See 3899-3902: Number of Positive and Examined Para-Aortic and Pelvic Nodes for additional
information
Coding Instructions and Codes
Note 1: Physician statement of positive para-aortic nodes can be used to code this data item when no
other information is available.
Note 2: The following are para-aortic nodes
Aortic
Lateral aortic/lateral lumbar
Para-aortic, NOS

290 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Periaortic
Note 3: Record the number of positive para-aortic lymph nodes documented in the medical record.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Micrometastasis and macrometastasis may be listed separately on the pathology report. Add
these two together to get the total number of positive nodes.
Note 6: Code X9 if no lymph node dissection is performed.
If only a FNA or core biopsy is done and it is positive, then code X6
If only a FNA or core biopsy is done and it is negative, then code X9
Code X9 when no lymph nodes are removed
Note 7: The number of examined para-aortic nodes is recorded in 3899: Number of Examined Para-
Aortic Nodes.
Code
Description
00 All para-aortic lymph nodes examined negative
01-
99
1-99 para-aortic lymph nodes positive
(Exact number of nodes positive)
X1 100 or more para-aortic nodes positive
X2 Positive para-aortic nodes identified, number unknown
X6 Positive aspiration or core biopsy of para-aortic lymph node(s)
X8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9 Not documented in medical record
Cannot be determined, indeterminate if positive para-aortic nodes present
No lymph node dissection performed
Para-aortic lymph nodes not assessed or unknown if assessed
Return to Schema ID Table

291 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3899: Number of Examined Para-Aortic Nodes
Item Length: 2
NAACCR Item #: 3899
XML Parent-NAACCR ID: Tumor-numberOfExaminedParaAorticNodes
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00528: Cervical Sarcoma (2021+)
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
00541: Corpus Sarcoma (2018+)
00542: Corpus Adenosarcoma (2018+)
Description
Number of Examined Para-Aortic nodes is the number of nodes examined based on para-aortic nodal
dissection.
Rationale
Number of Examined Para-Aortic Nodes is listed as a Registry Data Collection Variable in AJCC. This data
item was previously collected as Corpus, CS SSF #6.
Coding guidelines
Code 00 for when no nodes are examined
Code the exact number of examined nodes 01-99
Code X1 for 100 or more examined nodes
Code X2 for examined nodes, but unknown how many
Code X6 for aspiration or core biopsy of para-aortic node(s) only
Code X9 when
o Not documented in the medical record
o Para-Aortic lymph nodes not evaluated (assessed)
o No lymph node dissection performed
o Unknown if Para-Aortic lymph nodes not evaluated (assessed)
See 3899-3902: Number of Positive and Examined Para-Aortic and Pelvic Nodes for additional
information
Coding Instructions and Codes
Note 1: Physician statement of examined para-aortic nodes can be used to code this data item when no
other information is available.
Note 2: The following are para-aortic nodes
Aortic
Lateral aortic/lateral lumbar
Para-aortic, NOS

292 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Periaortic
Note 3: Record the number of examined para-aortic lymph nodes documented in the medical record.
Note 4: For the following
Code 00 when no lymph nodes are examined by FNA, core biopsy or removal of lymph node(s)
(e.g., sentinel lymph node biopsy or lymph node dissection)
o If a lymph node dissection is done and only pelvic lymph nodes are assessed, or only
“nodes” are documented without specifying pelvic or para-aortic, code to 00
Code X6 If only a FNA or core biopsy is done
Code X9 if it’s unknown if lymph nodes were removed
Note 5 : The number of positive para-aortic nodes is recorded in 3901: Number of Positive Para-Aortic
Nodes.
Code Description
00 No para-aortic nodes examined
01-99 1 - 99 para-aortic nodes examined
(Exact number of para-aortic lymph nodes examined)
X1 100 or more para-aortic nodes examined
X2 Para-aortic nodes examined, number unknown
X6
No para
-
aortic lymph nodes removed, but aspiration or core biopsy of
para
-
aortic node(s)
only
X8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9 Not documented in medical record
Cannot be determined, indeterminate if examined para-aortic nodes present
No lymph node dissection performed
Para-aortic lymph nodes not assessed or unknown if assessed
Return to Schema ID Table

293 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma
3902: Number of Positive Pelvic Nodes
Item Length: 2
NAACCR Item #: 3902
XML Parent-NAACCR ID: Tumor-numberOfPositivePelvicNodes
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00528: Cervical Sarcoma (2021+)
00530: Corpus Carcinoma and Carcinosarcoma
00541: Corpus Sarcoma
00542: Corpus Adenosarcoma
Description
Number of Positive Pelvic Nodes is the number of positive nodes based on pelvic nodal dissection.
Rationale
Number of Positive Pelvic Nodes is listed as a Registry Data Collection Variable in AJCC. This data item
was previously collected as Corpus, CS SSF #3.
See 3899-3902: Number of Positive and Examined Para-Aortic and Pelvic Nodes for additional
information
Coding guidelines
Code 00 for when there are no positive nodes
Code the exact number of positive nodes 01-99
Code X1 for 100 or more positive nodes
Code X2 for positive nodes, but unknown how many
Code X6 for aspiration or core biopsy of pelvic node(s) only
Code X9 when
o Not documented in the medical record
o Pelvic lymph nodes not evaluated (assessed)
o No lymph node dissection performed
o Unknown if Pelvic lymph nodes evaluated (assessed)
Additional Information
Source documents: pathology report, imaging reports, physical exam, other statements in
medical record
Coding Instructions and Codes
Note 1: Physician statement of positive pelvic nodes can be used to code this data item when no other
information is available.
Note 2: The following are pelvic nodes

294 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)
Paracervical
Parametrial
Pelvis, NOS
Sacral
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Presacral
o Uterosacral
Note 3: Record the number of positive pelvic lymph nodes documented in the medical record.
Note 4: For this data item, do not include isolated tumor cells (ITCs).
Note 5: Micrometastasis and macrometastasis may be listed separately on the pathology report. Add
these two together to get the total number of positive nodes.
Note 6: Code X9 if no lymph node dissection is performed.
If only a FNA or core biopsy is done and it is positive, then code X6
If only a FNA or core biopsy is done and it is negative, then code X9
Code X9 when no lymph nodes are removed
Note 7: The number of examined pelvic nodes is recorded in 3900: Number of Examined Pelvic Nodes.
Code
Description
00
All pelvic nodes examined negative
01-99 1 - 99 pelvic nodes positive
(Exact number of nodes positive)
X1 100 or more pelvic nodes positive
X2 Positive pelvic nodes identified, number unknown
X6 Positive aspiration or core biopsy of pelvic lymph node(s)
X8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9 Not documented in medical record
Cannot be determined, indeterminate if positive pelvic nodes present
No lymph node dissection performed
Pelvic lymph nodes not assessed or unknown if assessed
Return to Schema ID Table

295 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3900: Number of Examined Pelvic Nodes
Item Length: 2
NAACCR Item #: 3900
XML Parent-NAACCR ID: Tumor-numberOfExaminedPelvicNodes
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00528: Cervical Sarcoma (2021+)
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
00541: Corpus Sarcoma (2018+)
00542: Corpus Adenosarcoma (2018+)
Description
Number of Examined Pelvic Nodes is the number of nodes examined based on pelvic nodal dissection.
Rationale
Number of Examined Pelvic Nodes is listed as a Registry Data Collection Variable in AJCC. This data item
was previously collected as Corpus, CS SSF #4.
Coding guidelines
Code 00 for when no nodes are examined
Code the exact number of examined nodes 01-99
Code X1 for 100 or more examined nodes
Code X2 for nodes examined, but unknown how many
Code X6 for aspiration or core biopsy of pelvic(s) nodes only
Code X9 when
o Not documented in the medical record
o Pelvic lymph nodes not evaluated (assessed)
o No lymph node dissection performed
o Unknown if Pelvic lymph nodes not evaluated (assessed)
See 3899-3902: Number of Positive and Examined Para-Aortic and Pelvic Nodes for additional
information
Coding Instructions and Codes
Note 1: Physician statement of examined pelvic nodes can be used to code this data item when no other
information is available.
Note 2: The following are pelvic nodes
Iliac, NOS
o Common
o External
o Internal (hypogastric) (obturator)

296 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Paracervical
Parametrial
Pelvis, NOS
Sacral
o Lateral (laterosacral)
o Middle (promontorial) (Gerota’s node)
o Presacral
o Uterosacral
Note 3: Record the number of examined pelvic lymph nodes documented in the medical record.
Note 4: For the following
Code 00 when no lymph nodes are examined by FNA, core biopsy or removal of lymph node(s)
(e.g., sentinel lymph node biopsy or lymph node dissection)
o If a lymph node dissection is done and only “nodes” are documented without specifying
pelvic or para-aortic, assume they are pelvic
Code X6 If only a FNA or core biopsy is done
Code X9 if it’s unknown if lymph nodes were removed
Note 5: The number of positive pelvic nodes is recorded in 3902: Number of Positive Pelvic Nodes.
Code Description
00 No pelvic lymph nodes examined
01-
99
1 - 99 pelvic lymph nodes examined
(Exact number of pelvic lymph nodes examined)
X1 100 or more pelvic nodes examined
X2 Pelvic nodes examined, number unknown
X6 No pelvic lymph nodes removed, but aspiration or core biopsy of pelvic node(s) only
X8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9 Not documented in medical record
Cannot be determined, indeterminate if examined pelvic nodes present
No lymph node dissection performed
Pelvic lymph nodes not assessed or unknown if assessed
Return to Schema ID Table

297 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3911: Peritoneal Cytology
Item Length: 1
NAACCR Item #: 3911
XML Parent-NAACCR ID: Tumor-peritonealCytology
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00528: Cervical Sarcoma (2021+)
00530: Corpus Carcinoma and Carcinosarcoma (2018+)
00541: Corpus Sarcoma (2018+)
00542: Corpus Adenosarcoma (2018+)
Description
Peritoneal cytology pertains to the results of cytologic examination for malignant cells performed on
fluid that is obtained from the peritoneal cavity.
Rationale
Peritoneal Cytology is listed as a Registry Data Collection Variable in AJCC. This data item was previously
collected as Corpus, CS SSF #2.
Definition
Peritoneal cytology looks for malignant cells in the fluid in the pelvic and peritoneal cavities. Excess
natural fluid accumulation is called ascites. If, at laparotomy an analyzable amount of ascites is not
present, the surgeon may flood the pelvis and abdomen with saline solution then suction it out and send
the fluid for cytologic examination.
Additional Information
Source documents: cytology reports (look for multiple reports), pathology report
Other names: peritoneal washings, peritoneal lavage, possibly paracentesis (if no surgery)
Coding guidelines
Code 0 when the peritoneal cytology is reported as negative or normal
Code 1 when the peritoneal cytology test was done, and the results were reported as suspicious,
undetermined if negative or positive
Code 2 when the peritoneal cytology is reported as positive
Code 7 when test was ordered but the results are not in the medical record
Code 9 when
o No cytological specimen is available
o Peritoneal cytology not evaluated (assessed)
o Unknown if Peritoneal Cytology evaluated (assessed)

298 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note 1: Physician statement of Peritoneal Cytology can be used to code this data item when no other
information is available.
Note 2: Peritoneal cytology may also be called peritoneal ascitic fluid instead of peritoneal washing or
pelvic washing.
Note 3: Cytologic examination for malignant cells may be performed on ascites (fluid that has
accumulated in the peritoneal cavity in excess amount) or the fluid (saline) that is introduced into the
peritoneal cavity or pelvis, and then removed by suction. The introduction of fluid may be termed
peritoneal or pelvic washing or peritoneal lavage.
Code Description
0 Peritoneal cytology/washing negative for malignancy
1 Peritoneal cytology/washing atypical and/or suspicious
2 Peritoneal cytology/washing malignant (positive for malignancy)
3 Unsatisfactory/nondiagnostic
7 Test ordered, results not in chart
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9
Not documented in medical record
Peritoneal cytology not assessed or unknown if assessed
Return to Schema ID Table

299 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00541: Corpus Sarcoma (2018+)
See 00541: Corpus Sarcoma (2018+)
3836: FIGO Stage (Sarcoma)
See 00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3901: Number of Positive Para-Aortic Nodes
3899: Number of Examined Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3900: Number of Examined Pelvic Nodes
3911: Peritoneal Cytology
Return to Schema ID Table

300 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00541: Corpus Sarcoma (2018+)
3836: FIGO Stage (Sarcoma)
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2021+
Schema(s):
00528: Cervical Sarcoma (2021+)
00541: Corpus Sarcoma (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage
Code Description
1 FIGO Stage I
1A FIGO Stage IA
1B FIGO Stage IB
2 FIGO Stage II
2A FIGO Stage IIA
2B FIGO Stage IIB
3 FIGO Stage III
3A FIGO Stage IIIA
3B FIGO Stage IIIB
3C FIGO Stage IIIC
4
FIGO Stage IV
4A FIGO Stage IVA
4B FIGO Stage IVB
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
FIGO stage not assessed or unknown if assessed
Return to Schema ID Table

301 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00542: Corpus Adenosarcoma (2018+)
See 00541: Corpus Sarcoma (2018+)
3836: FIGO Stage (Sarcoma)
See 00530: Corpus Carcinoma and Carcinosarcoma (2018+)
3901: Number of Positive Para-Aortic Nodes
3899: Number of Examined Para-Aortic Nodes
3902: Number of Positive Pelvic Nodes
3900: Number of Examined Pelvic Nodes
3911: Peritoneal Cytology
Return to Schema ID Table

302 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00542: Corpus Adenosarcoma (2018+)
3836: FIGO Stage (Adenosarcoma)
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00542: Corpus Adenosarcoma (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Code Description
1 FIGO Stage I
1A FIGO Stage IA
1B FIGO Stage IB
1C FIGO Stage IC
2 FIGO Stage II
2A FIGO Stage IIA
2B
FIGO Stage IIB
3
FIGO Stage III
3A FIGO Stage IIIA
3B FIGO Stage IIIB
3C FIGO Stage IIIC
4 FIGO Stage IV
4A FIGO Stage IVA
4B FIGO Stage IVB
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

303 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00551: Ovary (2018+)
3836: FIGO: Ovary, Fallopian Tube, and Peritoneal Carcinoma
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00551: Ovary (2018+)
00552: Primary Peritoneal Carcinoma (2018+)
00553: Fallopian Tube (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Note 4: For High-grade (HGSC) serous tubal intraepithelial carcinoma (STIC) (8441/2), assign the FIGO
stage based on the managing physician's documentation of FIGO. (See Note 1).
If FIGO stage for HGSC or STIC is not documented by the managing physician, code unknown
(code 99)
Do not code 97 (in situ) for HGSC or STIC since FIGO does not have a Stage 0
If diagnosis is low grade serous intraepithelial carcinoma (LGSC) (8441/2) or serous
intraepithelial carcinoma (no grade stated) (8441/2), code 97
Note 5: Code 97 for any remaining in situ histologies (/2) since the FIGO stage definitions do not include
Stage 0.
Code Description
1 FIGO Stage I
1A FIGO Stage IA
1B FIGO Stage IB
1C FIGO Stage IC
1C1 FIGO Stage IC1
1C2 FIGO Stage IC2
1C3 FIGO Stage IC3
2 FIGO Stage II
2A FIGO Stage IIA
2B FIGO Stage IIB
3 FIGO Stage III

304 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
3A FIGO Stage IIIA
3A1 FIGO Stage IIIA1
3A11 FIGO Stage IIIA1i
3A12 FIGO Stage IIIA1ii
3A2 FIGO Stage IIIA2
3B
FIGO Stage IIIB
3C FIGO Stage IIIC
4 FIGO Stage IV
4A FIGO Stage IVA
4B FIGO Stage IVB
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
98
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99
Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

305 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00551: Ovary (2018+)
3818: CA-125 Pretreatment Interpretation
Item Length: 1
NAACCR Item #: 3818
XML Parent-NAACCR ID: Tumor-ca125PretreatmentInterpretation
NAACCR Alternate Name: CA-125 (Carbohydrate Antigen 125) Pretreatment Interpretation
Active years: 2018+
Schema(s):
00551: Ovary (2018+)
00552: Primary Peritoneal Carcinoma (2018+)
00553: Fallopian Tube (2018+)
Description
Carbohydrate Antigen 125 (CA-125)/CA-125 II is a tumor marker that is useful for following the response
to therapy in patients with ovarian cancer, who may have elevated levels of this marker.
Rationale
Preoperative CA-125/CA-125 II is a Registry Data Collection Variable listed in AJCC. It was previously
collected as Ovary, CS SSF #1.
Definition
CA-125/CA-125 II is a tumor marker that is not specific to ovarian or primary peritoneal cancer but is
useful to monitor for success of treatment and recurrence. Because it can be elevated in many diseases
affecting the peritoneal lining of the abdominal and pelvic cavity, it is not a screening test for women
who have no history of cancer. Any value over 35 is highly correlated with cancer and about 80% of
ovarian cancers show an elevated CA-125/CA-125 II. However, a result in the normal range does not
rule out cancer. Values up to 65 U/ml may be considered borderline, and values over 200 are unlikely to
be due to a benign condition. CA-125/CA-125 II monitors for success of treatment and
recurrence. After obtaining a baseline value prior to treatment, a lower result on a subsequent test
indicates a response to treatment, and an increasing value indicates possible recurrence.
Coding guidelines
Record the clinician’s interpretation of the highest value prior to treatment from a blood or serum test,
based on the reference range used by the lab. Do not code the result from thoracentesis or
paracentesis fluid.
Code 0 when the CA-125/CA-125 II is reported as negative or normal.
Code 1 when the CA-125/CA-125 II is reported as positive or elevated.
Code 2 when the CA-125/CA-125 II is reported as borderline; undetermined whether positive or
negative.
Code 7 when the CA-125/CA-125 II test was ordered but the results are not in the medical
record.
Code 9 when
o No information in the medical record
o CA-125/CA-125 II test not done (not assessed)

306 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Unknown if CA-125/CA-125 II test was performed (unknown if assessed)
Additional Information
Source documents: clinical laboratory report (blood or serum test); may be reported in history,
clinician or consultant notes or pathology report
Other names: Cancer Antigen 125, CA 125, CA125, Carbohydrate Antigen 125, mucin 16,
MUC16, CA 125 II
Normal reference range
o < 35 units per milliliter (U/ml); SI: < 35 kiloUnits/Liter (KU/L).
o May also be reported as micrograms/milliliter ( g/mL or ug/mL).
o Normal reference range may vary depending on the laboratory running the test.
Coding Instructions and Codes
Note 1: Physician statement of CA-125/CA-125 II pretreatment interpretation can be used to code this
data item when no other information is available.
Note 2: Carbohydrate Antigen 125 (CA-125/CA-125 II), also known as cancer antigen 125, mucin 16, or
MUC16, is a protein which in humans is encoded by the MUC16 gene. CA-125 is a tumor marker or
biomarker that may be elevated in the blood of some patients with ovarian cancer.
Note 3: Record only the blood or serum CA-125/CA-125 II interpretation for this data item. Do not
record CA-125 test results based on fluid from the chest or abdominal cavity.
Note 4: Record the CA-125/CA-125 II status prior to treatment.
Note 5: Normal values may vary with patient age and from lab to lab. The typical human reference
ranges are 0 to less than or equal 35 units per milliliter (U/mL). This is equivalent to kU/L.
Note 6: Code 9 if there is no statement that the CA-125/CA-125 II is positive/elevated, negative/normal,
and the lab value with its normal range (from which you can determine interpretation) is not
documented.
Code Description
0
Negative/normal; within normal limits
1
Positive/elevated
2 Stated as borderline; undetermined whether positive or negative
7 Test ordered, results not in chart
8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error)
9
Not documented in m
edical record
CA-125 not assessed or unknown if assessed
Return to Schema ID Table

307 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00551: Ovary (2018+)
3921: Residual Tumor Volume Post Cytoreduction
Item Length: 2
NAACCR Item #: 3921
XML Parent-NAACCR ID: Tumor-residualTumVolPostCytoreduction
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00551: Ovary (2018+)
00552: Primary Peritoneal Carcinoma (2018+)
00553: Fallopian Tube (2018+)
Description
Gross residual tumor after primary cytoreductive surgery is a prognostic factor for ovarian cancer and
residual tumor volume after cytoreductive surgery is a prognostic factor for late stage ovarian cancers.
Rationale
Residual Tumor Volume Post Cytoreduction is a Registry Data Collection Variable listed in AJCC. It was
previously collected as Ovary, CS SSF # 3.
Definition
The amount of ovarian tumor and the location of tumor remaining in the patient after initial ovarian or
peritoneal cancer surgery are the most important prognostic factors for advanced disease. The intent of
cytoreductive or debulking surgery—particularly for Stage III cancer—is to remove as much of the
cancer in the pelvis and abdomen as possible so that chemotherapy will be more effective. The less
tumor left behind, the more likely the patient will respond well to adjuvant hemotherapy. Information
about residual tumor volume will be in the operative report.
Additional Information
Source documents: operative report, discharge summary, chemotherapy records (inpatient and
outpatient)
For further information, refer to the Ovary, Fallopian Tube, or Peritoneum cancer protocol
published by the College of American Pathologists for the AJCC Staging System Ovary, Fallopian
Tube, and Primary Peritoneal Carcinoma
Other names: debulking, cytoreduction, residual tumor volume
Change for SSDI (effective v2.0): Further review of this SSDI indicated that the distinction of
whether patient had neoadjuvant therapy or not was not needed. The purpose of this data item
is to determine the residual tumor left behind. The codes have been redone so that they only
collect that information.

308 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note 1: Physician statement of residual tumor status after primary cytoreduction surgery can be used to
code this data item when no other information is available.
Note 2: Information for this SSDI is found in the operative report, procedure report, or managing
physician notes.
Note 3: The surgery to remove as much cancer in the pelvis and/or abdomen as possible, reducing the
"bulk" of the cancer, is called "debulking" or "cytoreductive" surgery. It is performed when there is
widespread evidence of advanced stage of ovarian cancer with obvious spread to other organs outside
the ovary, typically in the upper abdomen, intestines, the omentum (the fat pad suspended from the
transverse colon like an apron), the diaphragm, or liver.
Note 4: Optimal debulking is described as removal of all tumor except for residual nodules that measure
no more than 1 centimeter (cm) in maximum diameter.
Note 5: Gross residual tumor after primary cytoreductive surgery is a prognostic factor that has been
demonstrated in large studies. The best prognostic category after surgery includes those who are left
with no gross residual tumor.
Physicians should record the presence or absence of residual disease, if residual disease is
observed, the size of the largest visible lesion should be documented
Code Description
00 No gross residual tumor nodules
50 Residual tumor nodule(s) 1 centimeter (cm) or less
60 Residual tumor nodule(s) greater than 1 cm
70 Macroscopic residual tumor, size not stated
80 Procedure described as optimal debulking and size of residual tumor nodule(s) not given
97 No cytoreductive surgery performed
Non-invasive neoplasm (behavior /2)
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
Residual tumor status after cytoreductive surgery not assessed or unknown if assessed
Return to Schema ID Table

311 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00560: Placenta (2018+)
3836: FIGO: Gestational Trophoblastic Tumors (Placenta)
Item Length: 5
NAACCR Item #: 3836
XML Parent-NAACCR ID: Tumor-figoStage
NAACCR Alternate Name: FIGO Stage
Active years: 2018+
Schema(s):
00560: Placenta (2018+)
Note 1: There must be a statement about FIGO stage from the managing physician in order to code this
data item.
Do not code FIGO stage based on the pathology report
Do not code FIGO stage based only on T, N, M
If "FIGO" is not included with a stated stage, then do not assume it is a FIGO stage
Note 2: FIGO stage is not the same thing as FIGO grade. Only code FIGO stage in this field, do not code
FIGO grade.
Code FIGO grade in the grade fields
Note 3: If there is more than one FIGO stage provided from the clinical and pathological work up, code
the most extensive FIGO stage.
Note 4: The FIGO stage definitions do not include Stage 0 (Tis).
Code 97 for any non-invasive neoplasm (behavior /2)
Code
Description
1 FIGO Stage I
2 FIGO Stage II
3 FIGO Stage III
4 FIGO Stage IV
97 Not applicable: Carcinoma in situ (intraepithelial, noninvasive, preinvasive)
98 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 98 will result in an edit error.)
99 Not documented in medical record
FIGO stage unknown, not assessed or unknown if assessed
Return to Schema ID Table

312 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00560: Placenta (2018+)
3837: Gestational Trophoblastic Prognostic Scoring Index
Item Length: 2
NAACCR Item #: 3837
XML Parent-NAACCR ID: Tumor-gestationalTrophoblasticPxIndex
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00560: Placenta (2018+)
Description
Gestational Trophoblastic Prognostic Scoring Index, a score based on the FIGO-modified World Health
Organization (WHO) Prognostic Scoring Index, is used to stratify women with gestational trophoblastic
neoplasia in addition to the anatomical stage group. The risk score is appended to the anatomic stage.
Rationale
This data item is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 56 Gestational
Trophoblastic Neoplasms. It was previously collected as Placenta, CS SSF # 1.
Definition
The Prognostic Index is a non-anatomic risk factor scoring system that adds a fourth dimension to the
stage grouping of gestational trophoblastic tumors (GTT) of the placenta. The score subcategorizes GTTs
into low risk or high risk based on a point system. Code the clinician’s statement of the total point value
for the Prognostic Index in priority over the clinician’s statement of risk. Registrars are NOT to calculate
the score.
Coding Instructions and Codes
Note 1: This is based on clinician scoring only. The registrar is NOT to calculate the score based on
available information.
Note 2: The Prognostic Scoring Index is based on the following components
Age
Antecedent Pregnancy
Interval in Months from Index Pregnancy
Pretreatment Serum human chorionic gonadotropin (hCG) (mIU/ml)
Largest Tumor Size, Including Uterus
Sites of Metastases
Number of Metastases Identified
Previous Failed Chemotherapy
Note 3: The total score ranges from 00-25.
Note 4: If there is no clinician scoring, or a stated value is greater than 25, code X9.
314 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
MALE GENITAL ORGANS

316 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3920: PSA (Prostatic Specific Antigen) Lab Value
Item Length: 5
NAACCR Item #: 3920
XML Parent-NAACCR ID: Tumor-psaLabValue
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
PSA (Prostatic Specific Antigen) is a protein produced by cells of the prostate gland and is elevated in
patients with prostate cancer. This data item pertains to PSA lab value.
Rationale
This data item is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 58 Prostate. It was
previously collected as Prostate, CS SSF #1.
Definition
Serum PSA is the most sensitive tumor marker for monitoring individuals with prostate cancer, including
progression of disease and response to therapy. Although originally not intended to be a screening test,
this relatively simple blood test has become a very common method of detecting new prostate cancer in
its earliest stages. PSA can be totally negative when prostate cancer is found on digital rectal exam. In
such cases, PSA will not be helpful in monitoring for recurrence.
Note: Serum PSA is not the same as free PSA or precursor PSA—do not record values from
either of these tests in this field.
Additional Information
Source documents: clinical laboratory report (blood or serum test), history, clinician note,
pathology report
Other names: Prostate specific antigen, serum PSA, total PSA
Normal reference range: varies by age and race of patient.
The reference range should be shown on the clinical laboratory report. In general, normal
findings are 0 – 4.0 nanograms per milliliter (ng/ml).
Optimal normal range is 0 – 2.6 ng/ml. Nanograms per milliliter may be reported as micrograms
per liter ( g/L or ug/L).
Coding Guidelines
Record the last pre-diagnosis PSA lab value prior to diagnostic biopsy of prostate and initiation of
treatment in nanograms per milliliter (ng/ml) in the range 0.1 (.1 ng/ml) to 999.9 (999.9 ng/ml).

317 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note: This is a change from CSv2, where the instructions stated to code the highest PSA value
within 3 months prior to diagnostic biopsy
Examples Code Explanation
PSA 11.56 11.6 PSA documented in tenths, round up
1/5/2018: PSA 5.8
1/29/2018: PSA 5.2
2/22/2018: Biopsy positive for adenocarcinoma
5.2 PSA lab value closest and prior to the
diagnostic biopsy
12/19/2017: PSA 44.3
3/11/2018: PSA 42.8
5/1/2018: DRE positive for bilateral palpable
nodularity
5/5/2018: Casodex initiated without needle core
biopsy
42.8 PSA lab value closest to the initiation of
treatment
2/16/2018: PSA 18.6, adjusted PSA value due to
patient taking Medication for benign prostatic
hypertrophy
18.6 Record the adjusted PSA value ONLY if
documented by the clinician in the
record.
Registrar does not adjust the PSA value
due to BPH medication use
1,100 ng/ml XXX.1 XXX.1 is defined for values of 1,000 or
greater
No PSA done or unknown if done XXX.9 Definition of unknown
Coding Instructions and Codes
Note 1: Physician statement of prostatic specific antigen (PSA) pre-diagnosis can be used to code this
data item when no other information is available.
Note 2: PSA is a prognostic factor required for AJCC staging. It affects the stage group in most cases.
Note 3: Record to the nearest tenth in nanograms/milliliter (ng/ml) the last pre-diagnosis PSA lab value
prior to diagnostic biopsy of prostate and treatment. The lab value may be recorded in the lab report,
history and physical, or clinical statement in the pathology report, etc.
A lab value expressed in micrograms per liter (ug/L) is equivalent to the same value expressed in
nanograms per milliliter (ng/ml)
Record 0.1 when the lab results are stated as less than 0.1 ng/ml with no exact value.
Examples:
o PSA of 7.2. Code 7.2
o PSA of 10. Code 10.0
o PSA of 8.56. Code 8.6
o PSA of 110.35. Code 110.4
Note 4: A known lab value takes priority over codes XXX.2 and XXX.3
The lab value takes priority even if the physician documents the interpretation
Example: Patient noted to have a PSA of 7.6. Physician notes that the value is elevated
o Code 7.6 instead of XXX.3 (elevated)

318 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 5: A discrepancy between the PSA documented in the lab report and the PSA documented by the
clinician may arise due to the clinician’s adjusting the PSA value. Certain medications for benign
prostatic hypertrophy (BPH) decrease the PSA.
If there is documentation by a clinician within the medical record of an adjusted PSA value,
record the adjusted value.
The registrar does not adjust the PSA value based on BPH medication use.
If there is no documentation by a clinician within the medical record of an adjusted PSA value,
record the PSA value provided.
The fact that an adjusted PSA value is being recorded should be documented in the Dx Proc –
Lab Tests text field (NAACCR Item #2550).
Code Description
0.1 0.1 or less nanograms/milliliter (ng/ml)
(Exact value to nearest tenth of ng/ml)
0.2-999.9 0.2 – 999.9 ng/ml
(Exact value to nearest tenth of ng/ml)
XXX.1 1,000 ng/ml or greater
XXX.2 Lab value not available, physician states PSA is negative/normal
XXX.3
Lab value not available, physician states PSA is
positive/elevated/high
XXX.7 Test ordered, results not in chart
XXX.9 Not documented in medical record
PSA lab value not assessed or unknown if assessed
Return to Schema ID Table
319 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Prostate
Gleason Patterns and Scores
Definition
The Gleason system for grading prostate cancer is the one recommended by the AJCC and College of
American Pathologists. The following data items are used to collect information on Gleason.
3838: Gleason Patterns Clinical
3839: Gleason Patterns Pathological
3840: Gleason Score Clinical
3841: Gleason Score Pathological
3842: Gleason Tertiary Pattern
Gleason Patterns
The pathologist determines the Gleason patterns by looking at the prostate tissue under the
microscope. The pathologist assigns a grade to the most predominant pattern (largest surface area of
involvement, more than 50% of tissue) and a grade for the secondary pattern (second most
predominant) based on published Gleason criteria. When a patient undergoes radical prostatectomy,
the pathologist may look for a third or tertiary pattern in the specimen. When Gleason pattern 5 is
present as a tertiary pattern, its presence should be indicated in the pathology report, as a high Gleason
pattern appears to be an indicator for worse outcome (shortened time to recurrence). Studies indicate
that a Gleason score 7, with tertiary pattern 5, is associated with a worse prognosis than without tertiary
pattern 5 and is similar to the prognosis for Gleason score 8 – 10.
For example, in a specimen where the primary Gleason pattern is 3, the secondary is 4 and there
is less than 5% Gleason 5, the report should indicate a Gleason score of 7 (3+4) with tertiary
Gleason pattern 5. Gleason grades (patterns) range from 1 (small, uniform gland) to 5 (lack of
glands, sheets of cells.)
For the Gleason Patterns data items, there is a long list of codes and definitions in the table, but it may
be easier to assign a value if you understand the structure of the code. This is a two-digit field.
First digit is the Gleason primary pattern value
Second digit is the Gleason secondary pattern value
Gleason Score
The Gleason score is the sum of the values of the Gleason primary and secondary patterns. A low
Gleason score means the cancer tissue is similar to normal prostate tissue and the tumor is less likely to
spread; a high Gleason score means the cancer tissue is very different from normal and the tumor is
more likely to spread.
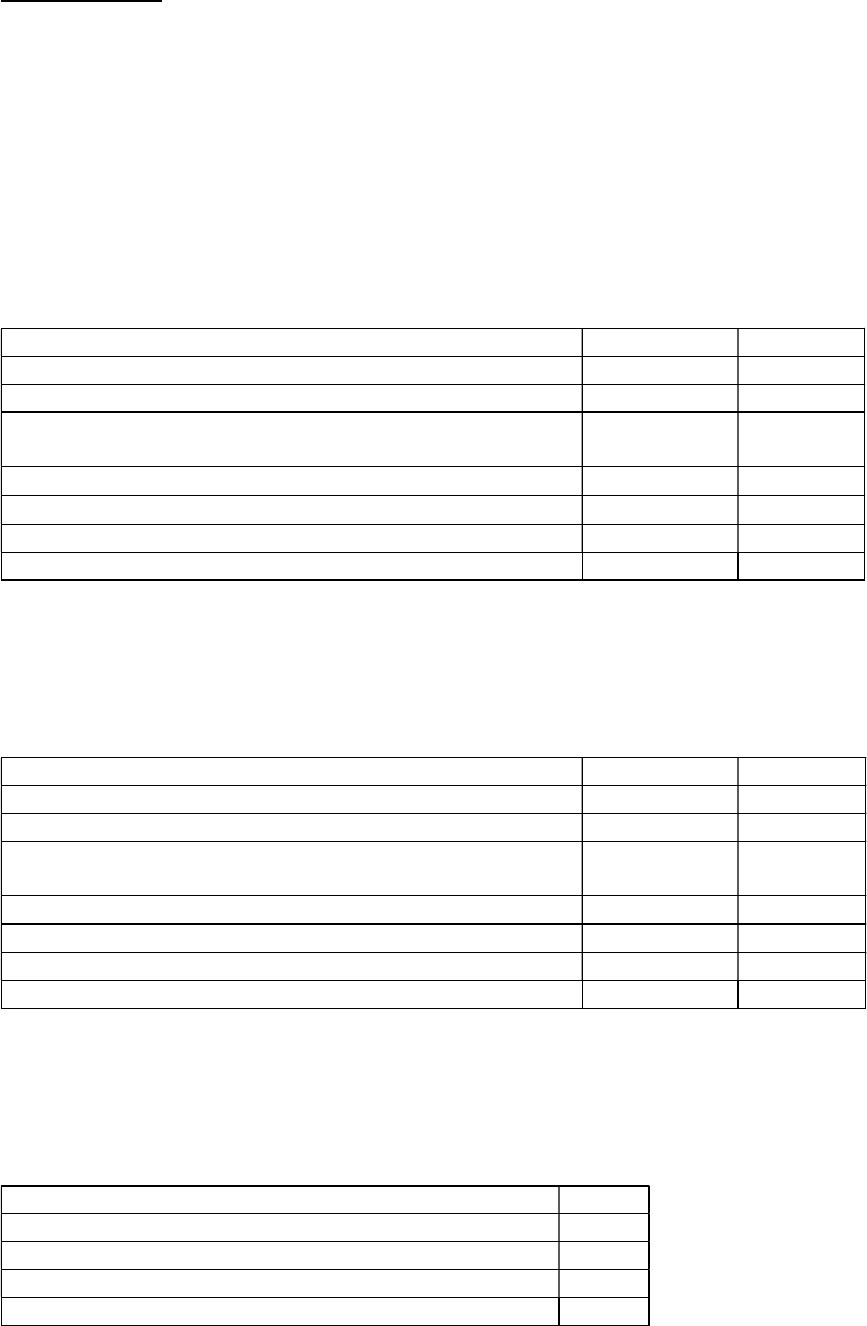
320 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding guidelines
Clinical Gleason Patterns and Score
Clinical Gleason Pattern and Score: Used to code information on the Gleason pattern from a needle core
biopsy or transurethral resection of the prostate (TURP) only. Gleason patterns from prostate tissue on a
transurethral resection of the bladder (TURB) specimen can also be used.
If there are multiple needle core biopsies or if both needle core biopsy and TURP are performed, code
the patterns and score from the specimen with the highest score.
Examples for Clinical Gleason Patterns and Score
Examples Pattern Code Score Code
Gleason 3+3 33 06
Gleason 4+3 43 07
Gleason 4 (Assume a number in the range 2-5 is a primary
pattern and code unknown (9) in the second digit)
49 X9
Gleason 7 (Assume a number in the range 6-10 is a score) X6 07
Gleason 10 (only combination of values that equals 10 is 5+5) 55 10
Needle core biopsy or TURP not done X7 X7
Gleason not done, or unknown if done X9 X9
Pathological Gleason Patterns and Score
Used to code information on the Gleason patterns from a prostatectomy or autopsy.
Examples for Pathological Gleason Patterns and Score
Examples Pattern Code Score Code
Gleason 3+3
33
06
Gleason 4+3
43
07
Gleason 4 (Assume a number in the range 2-5 is a primary
pattern and code unknown (9) in the second digit)
49 X9
Gleason 7 (Assume a number in the range 6-10 is a score) X6 07
Gleason 10 (only combination of values that equals 10 is 5+5) 55 10
No prostatectomy done X7 X7
Gleason not done, or unknown if done X9 X9
Tertiary Gleason Pattern
Used to code information on the Gleason tertiary pattern from a prostatectomy.
Examples for Tertiary Gleason Pattern
Examples Code
Tertiary pattern 3 30
Tertiary pattern 4 40
No prostatectomy done X7
Tertiary pattern not done, or unknown if done X9

321 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Clinical: pathology reports from needle biopsies or transurethral resection of prostate/bladder
that contains prostate tissue
o Pathological: pathology report from prostatectomy or autopsy report
For further information, refer to the Prostate cancer protocol published by the College of
American Pathologists for the AJCC Staging System Prostate
Return to Schema ID Table

322 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3838: Gleason Patterns Clinical
Item Length: 2
NAACCR Item #: 3838
XML Parent-NAACCR ID: Tumor-gleasonPatternsClinical
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
Prostate cancers are graded using Gleason score or pattern. This data item represents the Gleason
primary and secondary patterns from needle core biopsy or TURP.
Rationale
Gleason Patterns Clinical is a Registry Data Collection Variable for Clinical Stage for AJCC. This data item
was previously collected as Prostate, CS SSF #7.
See Gleason Patterns and Scores for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Gleason Patterns Clinical can be used to code this data item when there
is no other information available.
Note 2: Code the Gleason primary and secondary patterns from a needle core biopsy, trans rectal
ultrasound (TRUS) guided biopsy, or transurethral resection of prostate (TURP) and/or simple
prostatectomy in this field.
Gleason primary and secondary patterns provided for any prostate tissue identified from a
transurethral resection of a bladder tumor (TURBT) specimen can also be used in this field.
Note 3: Code the Gleason primary and secondary patterns prior to neoadjuvant treatment.
Note 4: Usually prostate cancers are graded using Gleason score or pattern. Gleason grading for
prostate primaries is based on a 5-component system (5 histologic patterns). Prostatic cancer generally
shows two main histologic patterns. The primary pattern, the pattern occupying greater than 50% of the
cancer, is usually indicated by the first number of the Gleason grade, and the secondary pattern is
usually indicated by the second number. These two numbers are added together to create a pattern
score.
If there are two numbers, assume that they refer to two patterns (the first number being the
primary pattern and the second number the secondary pattern), and sum them to obtain the
score.
If only one number is given, and it is less than or equal to 5, assume that it describes a pattern
(since scores of 5 or less would reflect Primary or Secondary Pattern Scores of 1 or 2). Code the
number as the primary pattern and code the secondary pattern as Unknown.
o For example, if only one number is given, and it is a 3, code “39” for Gleason Patterns
and “X9” for Gleason Score.
If only one number is given, and it is greater than 5, assume that it is a score.

323 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o For example, if only one number is given, and it is a 7, code “X6” for Gleason Patterns
and “07” for Gleason Score.
If the pathology report specifies a specific number out of a total of 10, the first number given is
the score.
o For example, if the pathology report says Gleason 7/10, code “07’ for Gleason Score and
“X6” for Gleason Patterns.
Note 5: If the only information available is the Gleason Score, code the patterns X6 (primary pattern
unknown, secondary pattern unknown).
Note 6: If different patterns are documented on multiple needle core biopsies, code the pattern that
reflects the highest or most aggressive score regardless if the pathologist provides an overall pattern in a
final summary. If different patterns equal the same high score, give priority to the highest primary
pattern and then the highest secondary pattern.
For example, both Gleason 3, 4 and Gleason 4, 3 equal Gleason score 7; code 43. Do not mix
patterns from multiple specimens.
Note 7: If multiple procedures are performed (e.g., needle core biopsy, trans rectal ultrasound (TRUS)
guided biopsy, transurethral resection of prostate (TURP), and/or simple prostatectomy), code the
pattern that reflects the highest score.
Note 8: Do not infer Gleason Primary and Secondary Pattern from Grade Group (Code X9).
Note 9: The clinical score is recorded in 3840: Gleason Score Clinical.
Code Description
11 Primary pattern 1, secondary pattern 1
12 Primary pattern 1, secondary pattern 2
13 Primary pattern 1, secondary pattern 3
14 Primary pattern 1, secondary pattern 4
15 Primary pattern 1, secondary pattern 5
19 Primary pattern 1, secondary pattern unknown
21 Primary pattern 2, secondary pattern 1
22 Primary pattern 2, secondary pattern 2
23
Primary pattern 2, secondary pattern 3
24 Primary pattern 2, secondary pattern 4
25 Primary pattern 2, secondary pattern 5
29 Primary pattern 2, secondary pattern unknown
31 Primary pattern 3, secondary pattern 1
32 Primary pattern 3, secondary pattern 2
33 Primary pattern 3, secondary pattern 3
34 Primary pattern 3, secondary pattern 4
35 Primary pattern 3, secondary pattern 5
39 Primary pattern 3, secondary pattern unknown
41 Primary pattern 4, secondary pattern 1
42 Primary pattern 4, secondary pattern 2
43 Primary pattern 4, secondary pattern 3
44 Primary pattern 4, secondary pattern 4
45 Primary pattern 4, secondary pattern 5
49
Primary pattern 4, secondary pattern unknown
51
Primary pattern 5, secondary pattern 1

324 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
52 Primary pattern 5, secondary pattern 2
53 Primary pattern 5, secondary pattern 3
54 Primary pattern 5, secondary pattern 4
55 Primary pattern 5, secondary pattern 5
59 Primary pattern 5, secondary pattern unknown
X6
TURP and/or Biopsy done, p
rimary pattern unknown, secondary pattern unknown
X7 No needle core biopsy/TURP performed
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit
error.)
X9
Not documented in medical record
Gleason Patterns Clinical not assessed or unknown if assessed
Unknown whether TURP and/or Biopsy done
Return to Schema ID Table

325 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3840: Gleason Score Clinical
Item Length: 2
NAACCR Item #: 3840
XML Parent-NAACCR ID: Tumor-gleasonScoreClinical
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
This data item records the Gleason score based on adding the values for primary and secondary patterns
in Needle Core Biopsy or TURP.
Rationale
Gleason Score Clinical is a Registry Data Collection Variable for AJCC. This data item was previously
collected as Prostate, CS SSF #8.
See Gleason Patterns and Scores for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Gleason Score Clinical can be used to code this data item when there is
no other information available.
Note 2: Code the Gleason Score Clinical from a needle core biopsy, trans rectal ultrasound (TRUS)
guided biopsy, transurethral resection of prostate (TURP), and/or simple prostatectomy in this field.
Gleason primary and secondary patterns provided for any prostate tissue identified from a
transurethral resection of a bladder tumor (TURBT) specimen can also be used in this field.
Note 3: Code the Gleason Score prior to neoadjuvant treatment.
Note 4: Usually prostate cancers are graded using Gleason's score or pattern. Gleason's grading for
prostate primaries is based on a 5-component system (5 histologic patterns). Prostatic cancer generally
shows two main histologic patterns. The primary pattern, the pattern occupying greater than 50% of the
cancer, is usually indicated by the first number of the Gleason's grade, and the secondary pattern is
usually indicated by the second number. These two numbers are added together to create a pattern
score, ranging from 2 to 10.
If there are two numbers, assume that they refer to two patterns (the first number being the
primary pattern and the second number the secondary pattern), and sum them to obtain the
score.
If only one number is given, and it is less than or equal to 5, code the total score to X9, unknown
or no information.
If only one number is given, and it is greater than 5, assume that it is a score and code as stated.
If the pathology report specifies a specific number out of a total of 10, the first number given is
the score.

326 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o Example: The pathology report says Gleason's 3/10. The Gleason's score would be 3 and
coded as 03.
Note 5: If different scores are documented on multiple needle core biopsies, code the highest or most
aggressive score.
Note 6: If multiple procedures are performed (e.g., needle core biopsy, trans rectal ultrasound (TRUS)
guided biopsy, transurethral resection of prostate (TURP), and/or simple prostatectomy), code the
highest score.
Note 7: Do not infer the Gleason Score from Grade Group (Code X9).
Note 8: Record the Gleason score based on the addition of the primary and secondary patterns coded in
3838: Gleason Patterns Clinical.
Code Description
02 Gleason score 2
03 Gleason score 3
04 Gleason score 4
05 Gleason score 5
06 Gleason score 6
07
Gleason score 7
08
Gleason score 8
09 Gleason score 9
10 Gleason score 10
X7 No needle core biopsy/TURP performed
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit
error.)
X9 Not documented in medical record
Gleason Score Clinical not assessed or unknown if assessed
Return to Schema ID Table

327 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3839: Gleason Patterns Pathological
Item Length: 2
NAACCR Item #: 3839
XML Parent-NAACCR ID: Tumor-gleasonPatternsPathological
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
Prostate cancers are graded using Gleason score or pattern. This data item represents the Gleason
primary and secondary patterns from prostatectomy or autopsy.
Rationale
Gleason Patterns Pathological is a Registry Data Collection Variable for AJCC. This data item was
previously collected as Prostate, CS SSF #9.
See Gleason Patterns and Scores for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Gleason Patterns Pathological can be used to code this data item when
there is no other information available.
Note 2: Code the Gleason primary and secondary patterns from a radical prostatectomy or autopsy only
in this field. Unlike Grade Group Pathological, do not include patterns from tissues taken prior to a
radical prostatectomy.
Code results from a transurethral resection of prostate (TURP) or simple prostatectomy in
Gleason Patterns Clinical
Note 3: Usually prostate cancers are graded using Gleason score or pattern. Gleason grading for
prostate primaries is based on a 5-component system (5 histologic patterns). Prostatic cancer generally
shows two main histologic patterns. The primary pattern, the pattern occupying greater than 50% of the
cancer, is usually indicated by the first number of the Gleason grade, and the secondary pattern is
usually indicated by the second number. These two numbers are added together to create a pattern
score.
If there are two numbers, assume that they refer to two patterns (the first number being the
primary pattern and the second number the secondary pattern), and sum them to obtain the
score.
If only one number is given, and it is less than or equal to 5, assume that it describes a pattern
(since scores of 5 or less would reflect Primary or Secondary Pattern Scores of 1 or 2). Code the
number as the primary pattern and code the secondary pattern as Unknown.
o For example, if only one number is given, and it is a 3, code “39” for Gleason Patterns
and “X9” for Gleason Score.
If only one number is given, and it is greater than 5, assume that it is a score.

328 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o For example, if only one number is given, and it is a 7, code “X6” for Gleason Patterns
and “07” for Gleason Score.
If the pathology report specifies a specific number out of a total of 10, the first number given is
the score.
o For example, if the pathology report says Gleason 7/10, code “07” for Gleason Score
and “X6” for Gleason Patterns.
Note 4: If the only information available is the Gleason Score, code the patterns X6 (primary pattern
unknown, secondary pattern unknown).
Note 5: If different patterns are documented on multiple specimens, code the pattern that reflects the
highest or most aggressive score regardless if the pathologist provides and overall pattern in a final
summary. If different patterns equal the same high score, give priority to the highest primary pattern
and then the highest secondary pattern.
Note 6: If neoadjuvant therapy was given, code Gleason pathological patterns as X9.
Note 7: Do not infer Gleason Primary and Secondary Pattern from Grade Group (Code X9).
Note 8: If a tertiary pattern is documented on prostatectomy or autopsy, code in 3842: Gleason Tertiary
Pattern.
Note 9: The pathological score is recorded in 3841: Gleason Score Pathological.
Code Description
11 Primary pattern 1, secondary pattern 1
12 Primary pattern 1, secondary pattern 2
13
Primary pattern 1, secondary pattern 3
14
Primary pattern 1, secondary pattern 4
15 Primary pattern 1, secondary pattern 5
19 Primary pattern 1, secondary pattern unknown
21 Primary pattern 2, secondary pattern 1
22 Primary pattern 2, secondary pattern 2
23 Primary pattern 2, secondary pattern 3
24 Primary pattern 2, secondary pattern 4
25 Primary pattern 2, secondary pattern 5
29 Primary pattern 2, secondary pattern unknown
31 Primary pattern 3, secondary pattern 1
32 Primary pattern 3, secondary pattern 2
33 Primary pattern 3, secondary pattern 3
34 Primary pattern 3, secondary pattern 4
35 Primary pattern 3, secondary pattern 5
39 Primary pattern 3, secondary pattern unknown
41
Primary pattern 4, secondary pattern 1
42
Primary pattern 4, secondary pattern 2
43 Primary pattern 4, secondary pattern 3
44 Primary pattern 4, secondary pattern 4
45 Primary pattern 4, secondary pattern 5
49 Primary pattern 4, secondary pattern unknown
51 Primary pattern 5, secondary pattern 1
52 Primary pattern 5, secondary pattern 2

329 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
53 Primary pattern 5, secondary pattern 3
54 Primary pattern 5, secondary pattern 4
55 Primary pattern 5, secondary pattern 5
59 Primary pattern 5, secondary pattern unknown
X6 Prostatectomy done, primary pattern unknown, secondary pattern unknown
X7
No
radical
prostatectomy/autopsy performed
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit
error.)
X9 Not documented in medical record
Gleason Patterns Pathological not assessed or unknown if assessed
Unknown if radical prostatectomy done
Return to Schema ID Table

330 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3841: Gleason Score Pathological
Item Length: 2
NAACCR Item #: 3841
XML Parent-NAACCR ID: Tumor-gleasonScorePathological
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
This data item records the Gleason score based on adding the values for primary and secondary patterns
from prostatectomy or autopsy.
Rationale
Gleason Score Pathological is a Registry Data Collection Variable for AJCC. This data item was previously
collected as Prostate, CS SSF #10.
See Gleason Patterns and Scores for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Gleason Score Pathological can be used to code this data item when
there is no other information available.
Note 2: Code the Gleason Score Pathological from a radical prostatectomy or autopsy only in this field.
Unlike Grade Group Pathological, do not include patterns from tissues taken prior to a radical
prostatectomy.
Code results from a transurethral resection of prostate (TURP) or simple prostatectomy in
Gleason Score Clinical
Note 3: Usually prostate cancers are graded using Gleason's score or pattern. Gleason's grading for
prostate primaries is based on a 5-component system (5 histologic patterns). Prostatic cancer generally
shows two main histologic patterns. The primary pattern, the pattern occupying greater than 50% of the
cancer, is usually indicated by the first number of the Gleason's grade, and the secondary pattern is
usually indicated by the second number. These two numbers are added together to create a pattern
score, ranging from 2 to 10.
If there are two numbers, assume that they refer to two patterns (the first number being the
primary pattern and the second number the secondary pattern), and sum them to obtain the
score.
If only one number is given, and it is less than or equal to 5, code the total score to X9, unknown
or no information.
If only one number is given, and it is greater than 5, assume that it is a score and code as stated.
If the pathology report specifies a specific number out of a total of 10, the first number given is
the score.
o Example: The pathology report says Gleason's 3/10. The Gleason's score would be 3 and
coded as 03.

331 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: If neoadjuvant therapy was given, code Gleason pathological score as X9.
Note 5: Do not infer the Gleason Score from Grade Group (Code X9).
Note 6: Record the Gleason score based on the addition of the primary and secondary patterns coded in
3839: Gleason Patterns Pathological.
Code Description
02 Gleason score 2
03 Gleason score 3
04 Gleason score 4
05 Gleason score 5
06 Gleason score 6
07 Gleason score 7
08 Gleason score 8
09 Gleason score 9
10 Gleason score 10
X7 No radical prostatectomy done/autopsy performed
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit
error.)
X9 Not documented in medical record
Gleason Score Pathological not assessed or unknown if assessed
Unknown if radical prostatectomy done
Return to Schema ID Table

332 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3842: Gleason Tertiary Pattern
Item Length: 2
NAACCR Item #: 3842
XML Parent-NAACCR ID: Tumor-gleasonTertiaryPattern
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
Prostate cancers are graded using Gleason score or pattern. This data item represents the tertiary
pattern value from prostatectomy or autopsy.
Rationale
Tertiary Gleason pattern on prostatectomy is a Registry Data Collection Variable for AJCC. This data item
was previously collected as Prostate, CS SSF #11.
See Gleason Patterns and Scores for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Gleason tertiary pattern can be used to code this data item when there is
no other information available.
Note 2: If present, a high Gleason Tertiary Pattern appears to be an indication for a worse outcome.
Note 3: Record the tertiary pattern documented on radical prostatectomy or autopsy only. Record the
tertiary pattern prior to neoadjuvant treatment.
If a tertiary pattern is documented on needle core biopsy or transurethral resection of prostate
(TURP), it should be disregarded
Do not code the tertiary pattern on radical prostatectomy or autopsy in Gleason Patterns
Pathological
Note 4: The CAP Prostate protocol does not include Patterns 1 and 2 for Tertiary Pattern.
Note 5: If neoadjuvant therapy was given, code Gleason patterns as X9.
Code Description
10
Tertiary pattern 1
20
Tertiary pattern 2
30 Tertiary pattern 3
40 Tertiary pattern 4
50 Tertiary pattern 5
X7 No radical prostatectomy/autopsy performed
X8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit error.)
X9
Not documented in medical record
Gleason Tertiary Pattern not assessed or unknown if assessed
Return to Schema ID Table

333 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
Number of Cores Positive and Examined
Definition
These two data items record the number of positive and examined cores that are microscopically
confirmed. A diagnostic procedure, such as a needle core biopsy, can take as many as 20 or more core
biopsies to determine the extent of the cancer within the prostate.
Together these two data items can provide researchers with a surrogate estimate of the percentage of
the prostate involved by tumor, if that figure is not stated in the pathology report
Number of Cores Positive must ALWAYS be less than or equal to Number of Cores Examined.
For Prostate, there are 2 data items that record information on the number of cores positive and
examined. These data items should be coded from the same test.
3897: Number of Cores Examined
3898: Number of Cores Positive
Note: Do not make assumptions about the number of cores positive or examined based on the
number of areas biopsied within the prostate (laterality, lobes, apex, base, or mid-prostate). Several
cores may be taken from each area.
Additional Information
Clinical: pathology reports from needle biopsies or transurethral resection of prostate/bladder
that contains prostate tissue
For further information, refer to the Prostate cancer protocol published by the College of
American Pathologists for the AJCC Staging System Prostate
Source documents: pathology reports from core needle biopsies
Other names for procedures: needle core biopsy, needle biopsy, core biopsy, prostate biopsy,
sextant biopsy, transrectal biopsy, ultrasound-guided biopsy, transperineal prostate biopsy,
triggered-needle biopsy.
Return to Schema ID Table

334 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3898: Number of Cores Positive
Item Length: 2
NAACCR Item #: 3898
XML Parent-NAACCR ID: Tumor-numberOfCoresPositive
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
This data item represents the number of positive cores documented in the pathology report from
needle biopsy of the prostate gland.
Rationale
Number of Cores Positive is a Registry Data Collection Variable for AJCC. This data item was previously
collected as Prostate, CS SSF #12.
Coding guidelines
Code 00 for all cores negative
Code the exact number of positive cores 01-99
Code X1 for 100 or more positive cores
Code X6 for positive cores, unknown how many
Code X9 when
o Not documented in the medical record
o Cores not evaluated (assessed)
o Unknown if Cores evaluated (assessed)
See Number of Cores Positive and Examined for additional information.
Coding Instructions and Codes
Note 1: Physician statement of Number of Cores Positive can be used to code this data item when there
is no other information available.
Note 2: Record the number of positive prostate core biopsies from the first prostate core biopsy
diagnostic for cancer. If positive cores are identified and the number of positive cores not specifically
documented, code X6.
Information from the first core biopsy is preferred since the physician is usually examining the
entire prostate. If a second core biopsy is done, this is usually done on a specified area, so more
cores will be found to be positive
Note 3: If the pathology report contains a summary of the number of cores positive and examined, use
the summary provided. If Summary Report is not available and multiple biopsy cores are obtained on
the same day, the number of cores examined should be added.

335 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Do not include cores of other area like seminal vesicles
Information from the gross description of the core biopsy pathology report can be used to code
this data item when the gross findings provide the actual number of cores and not pieces, chips,
fragments, etc.
Note 4: Transperineal template-guided saturation biopsy (TTSB) is a stereotactic prostate biopsy
technique that typically produces 30 to 80 core biopsies. This is an alternative biopsy technique used for
some high-risk patients including men with persistently elevated PSA, those who have atypia on prior
prostate biopsies, or men with biopsies showing high grade prostatic intraepithelial neoplasia (PIN).
Note 5: The number of cores examined is recorded in 3897: Number of Cores Examined.
Code Description
00 All examined cores negative
01-99 1 - 99 cores positive
(Exact number of cores positive)
X1 100 or more cores positive
X6 Biopsy cores positive, number unknown
X7 No needle core biopsy performed
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit
error.)
X9
Not documented in medical record
Number of Cores Positive not assessed or unknown if assessed
Return to Schema ID Table

336 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00580: Prostate (2018+)
3897: Number of Cores Examined
Item Length: 2
NAACCR Item #: 3897
XML Parent-NAACCR ID: Tumor-numberOfCoresExamined
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00580: Prostate (2018+)
Description
This data item represents the number of cores examined as documented in the pathology report from
needle biopsy of the prostate gland.
Rationale
Number of Cores Examined is a Registry Data Collection Variable for AJCC. This data item was previously
collected as Prostate, CS SSF #13.
Coding guidelines
Code the exact number of examined cores 01-99
Code X1 for 100 or more examined cores
Code X6 for examined cores, unknown how many
Code X9 when
o Not documented in the medical record
o Cores not evaluated (assessed)
o Unknown if Cores evaluated (assessed)
See Number of Cores Positive and Examined for additional information.
Coding Instructions and Notes
Note 1: Physician statement of Number of Cores Examined can be used to code this data item when
there is no other information available.
Note 2: Record the number of prostate core biopsies examined from the first prostate core biopsy
diagnostic for cancer. If the number of cores examined is not specifically documented, code X6.
Information from the first core biopsy is preferred since the physician is usually examining the
entire prostate. If a second core biopsy is done, this is usually done on a specified area, so more
cores will be found to be positive
Note 3: If the pathology report contains a summary of the number of cores positive and examined, use
the summary provided. If Summary Report is not available and multiple biopsy cores are obtained on
the same day, the number of cores examined should be added.
Do not include cores of other area like seminal vesicles

337 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Information from the gross description of the core biopsy pathology report can be used to code
this data item when the gross findings provide the actual number of cores and not pieces, chips,
fragments, etc.
Note 4: Transperineal template-guided saturation biopsy (TTSB) is a stereotactic prostate biopsy
technique that typically produces 30 to 80 core biopsies. This is an alternative biopsy technique used for
some high-risk patients including men with persistently elevated PSA, those who have atypia on prior
prostate biopsies, or men with biopsies showing high grade prostatic intraepithelial neoplasia (PIN).
Note 5: The number of cores positive is recorded in 3898: Number of Cores Positive.
Code
Description
01
-
99
1
-
99 cores
examined
(Exact number of cores examined)
X1
100 or more cores examined
X6
Biopsy cores examined, number unknown
X7 No needle core biopsy performed
X8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code X8 may result in an edit
error.)
X9
Not documented in medical record
Number of cores examined not assessed or unknown if assessed
Return to Schema ID Table

338 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
Testis Serum Markers and S Category
In addition to T, N, and M, the S category is collected to stage Testicular cancers. There are three factors
that make up the S stage: alpha-fetoprotein (AFP), beta-human chorionic gonadotropin (beta-hCG), and
lactase dehydrogenase (LDH). These play an important role as serum tumor markers in the staging and
monitoring of germ cell tumors and should be measured prior to removing the involved testicle. For
patients with nonseminomas, the degree of tumor-marker elevation after the cancerous testicular has
been removed is one of the most significant predictors of prognosis. Serum tumor markers are also very
useful for monitoring all stages of nonseminomas and for monitoring metastatic seminomas because
elevated marker levels are often the earliest sign of relapse.
There are several data items related to the collection of these variables.
For clinical staging
3807: AFP Pre-Orchiectomy Lab Value
3808: AFP Pre-Orchiectomy Range
3848: hCG Pre-Orchiectomy Lab Value
3849: hCG Pre-Orchiectomy Range
3868: LDH Pre-Orchiectomy Range
3923: S Category Clinical
For pathological staging
3805: AFP Post-Orchiectomy Lab Value
3806: AFP Post-Orchiectomy Range
3846: hCG Post-Orchiectomy Lab Value
3847: hCG Post-Orchiectomy Range
3867: LDH Post-Orchiectomy Range
3924: S Category Pathological
In Collaborative Stage v2 (CSv2), the “ranges” were used to derive the S category. New data items for
AJCC 8
th
edition is for the assignment of the S category in addition to collecting the individual data items.
Return to Schema ID Table
339 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
Alpha-fetoprotein (AFP) (Testis)
Definition
Alpha-fetoprotein (AFP) is a protein normally made by immature liver cells in the fetus. In adults, high
AFP levels (> 500 ng/ml) in the blood occur only in hepatocellular carcinoma (>1000), liver metastases
(from a primary elsewhere), and germ cell tumors of the testes and ovaries. Elevated AFP values are
found in non-seminomatous malignancies and mixed tumors of the testis. AFP is used with HCG to
identify the specific cell type of testicular cancer. AFP is not secreted by pure seminoma or teratoma. If
AFP > 500 ng/ml, the underlying condition is unlikely to be benign. If AFP > 10,000 ng/ml at diagnosis,
the patient is likely to have a poor prognosis.
AFP is more useful in monitoring response to therapy than making a diagnosis. The half-life of AFP is 5 to
7 days. After orchiectomy, the AFP should fall to < 25 ng/ml in 25-35 days. If elevated AFP persists, this is
an indication of residual tumor.
For Testis, there are 4 data items that record information on AFP for Testis.
3805: AFP Post-Orchiectomy Lab Value
3806: AFP Post-Orchiectomy Range
3807: AFP Pre-Orchiectomy Lab Value
3808: AFP Pre-Orchiectomy Range
Coding guidelines
Assign the code for the highest AFP value and corresponding AFP range prior to orchiectomy. In the
event an orchiectomy is not performed, or systemic treatment precedes an orchiectomy, code the
highest AFP value and corresponding range prior to any systemic treatment. The AFP Range is a category
used to group the lab values into 3 ranges: 1, 2, and 3. The pre-orchiectomy AFP lab value and the pre-
orchiectomy AFP range should be from the same test. If the clinician states an S value rather than a lab
value, code unknown (code XXXX.9) for the AFP pre-orchiectomy lab value and unknown (code 9) for the
AFP pre-orchiectomy range.
Categories used for Pre- and Post-Orchiectomy AFP Range are:
Code Description
0 Within normal limits
1 Above normal and less than 1,000 nanograms/milliliter (ng/mL)
2 1,000 -10,000 ng/mL
3 Greater than 10,000 ng/mL

340 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Examples for AFP Pre-Orchiectomy and AFP Post-Orchiectomy Lab Value and Range
For these examples, the lab’s normal reference range for AFP = 0-10 ng/ml
Examples Lab Value Code Range Code
1 ng/ml 1.0 0
270 ug/l 270.0
(ng/ml = ug/L)
1
5500 ng/ml 5500.0 2
12,500 ng/ml 12500.0 3
110,000 ng/ml XXXXX.1 3
Physician states “AFP elevated,” but no value
documented
XXXXX.9 4
S value stated (no other information available) XXXXX.9 9
No AFP test done, or unknown if done XXXXX.9 9
Additional Information
o Source documents: clinical laboratory report (blood serum radioimmunoassay or enzyme assay
(EIA)); sometimes in history and physical or clinical statement in pathology report
o For further information, refer to the Testis cancer protocol published by the College of American
Pathologists for the AJCC Staging System Testis
o Other names: FP, aFP, Alpha Fetoprotein, Alpha-fetoprotein, -fetoprotein; fetal alpha
globulin
o Normal Reference Range: Adult men 0-15 ng/ml (SI: 0-15 µg/L)
o Measurements: micrograms/liter (µg/L or ug/L) is equivalent to nanograms per milliliter (ng/ml)
o If measurements are given in IU/ml, use the following conversion:
1 ng/mL = 0.83 IU/mL
To calculate ng from IU/mL, divide the value for IU by 0.83.
Example: 10 IU/mL: 10/0.83 = 12.04 ng/mL; 5 IU/mL: 5/0.83= 6.02
ng/mL
Return to Schema ID Table

341 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3807: AFP Pre-Orchiectomy Lab Value
Item Length: 7
NAACCR Item #: 3807
XML Parent-NAACCR ID: Tumor-afpPreOrchiectomyLabValue
NAACCR Alternate Name: AFP (Alpha Fetoprotein) Pre-Orchiectomy Lab Value
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
AFP (Alpha Fetoprotein) Pre-Orchiectomy Lab Value refers to the AFP value measured prior to
treatment. AFP is a tumor marker that is often elevated in patients with nonseminomatous germ cell
tumors of the testis.
Rationale
AFP (Alpha Fetoprotein) Pre-Orchiectomy Lab Value is a Registry Data Collection Variable in AJCC. It was
previously collected as Testis CS SSF #6.
See Alpha-fetoprotein (AFP) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the AFP (Alpha Fetoprotein) Pre-Orchiectomy Lab Value can be used to
code this data item when no other information is available.
Note 2: Record the lab value of the highest AFP test result documented in the medical record prior to
orchiectomy or prior to any systemic treatment. The lab value may be documented in a lab report,
history and physical, or clinical statement in the pathology report.
Note 3: A lab value expressed in micrograms/liter (ug/l) is equivalent to the same value expressed in
ng/mL.
Note 4: If the lab value is expressed in IU/ml, use the following conversion: 1 ng/mL = 0.83 IU/mL.
o To calculate ng from IU/mL, divide the value for IU by 0.83.
o Example: 10 IU/mL: 10/0.83 = 12.04 ng/mL; 5 IU/mL: 5/0.83= 6.02 ng/mL
Note 5: The same laboratory test should be used to record information in 3808: AFP Pre-Orchiectomy
Range.
Code Description
0.0 0.0 nanograms/milliliter (ng/mL)
0.1-
99999.9
0.1 – 99,999.9 ng/mL
XXXXX.1 100,000 ng/mL or greater
XXXXX.7 Test ordered, results not in chart

342 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
XXXXX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XXXXX.8 may result
in an edit error.)
XXXXX.9 Not documented in medical record
AFP (Alpha Fetoprotein) Pre-Orchiectomy Lab Value not assessed or unknown if
assessed
Return to Schema ID Table

343 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3808: AFP Pre-Orchiectomy Range
Item Length: 1
NAACCR Item #: 3808
XML Parent-NAACCR ID: Tumor-afpPreOrchiectomyRange
NAACCR Alternate Name: AFP (Alpha Fetoprotein) Pre-Orchiectomy Range
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
AFP (Alpha Fetoprotein) Pre-Orchiectomy Range identifies the range category of the highest AFP value
measured prior to treatment. AFP is a serum tumor marker that is often elevated in patients with
nonseminomatous germ cell tumors of the testis.
Rationale
AFP (Alpha Fetoprotein) is a Registry Data Collection Variable in AJCC. AFP (Alpha Fetoprotein) Pre-
Orchiectomy Range is used to assign the S Category Clinical and was previously collected as Testis CS SSF
#7.
See Alpha-fetoprotein (AFP) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the AFP (Alpha Fetoprotein) Pre-Orchiectomy Range can be used to code
this data item when no other information is available.
Note 2: Record the range of the highest AFP test result documented in the medical record prior to
orchiectomy or prior to any systemic treatment. The lab value may be documented in a lab report,
history and physical, or clinical statement in the pathology report.
Note 3: A lab value expressed in micrograms/liter (ug/L) is equivalent to the same value expressed in
nanograms/milliliter (ng/mL).
Note 4: If the lab value is expressed in IU/ml, use the following conversion: 1 ng/mL = 0.83 IU/mL.
o To calculate ng from IU/mL, divide the value for IU by 0.83.
o Example: 10 IU/mL: 10/0.83 = 12.04 ng/mL; 5 IU/mL: 5/0.83= 6.02 ng/mL
Note 5: The same laboratory test should be used to record information in 3807: AFP Pre-Orchiectomy
Lab Value.
Code Description
0
Within normal limits
1
Above normal and less than 1,000 nanograms/milliliter (ng/mL)
2 1,000 -10,000 ng/mL
3 Greater than 10,000 ng/mL

344 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
4 Pre-Orchiectomy alpha fetoprotein (AFP) stated to be elevated
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
AFP (Alpha Fetoprotein) Pre-Orchiectomy Range not assessed or unknown if assessed
Return to Schema ID Table

345 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3805: AFP Post-Orchiectomy Lab Value
Item Length: 7
NAACCR Item #: 3805
XML Parent-NAACCR ID: Tumor-afpPostOrchiectomyLabValue
NAACCR Alternate Name: AFP (Alpha Fetoprotein) Post-Orchiectomy Lab Value
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
AFP (Alpha Fetoprotein) Post-Orchiectomy Lab Value refers to the lowest AFP value measured post-
orchiectomy. AFP is a serum tumor marker that is often elevated in patients with nonseminomatous
germ cell tumors of the testis. The Post-Orchiectomy lab value is used to monitor response to therapy.
Rationale
AFP (Alpha Fetoprotein) Post-Orchiectomy Lab Value is a Registry Data Collection Variable in AJCC. It
was previously collected as Testis CS SSF #12.
See Alpha-fetoprotein (AFP) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the AFP (Alpha Fetoprotein) Post-Orchiectomy Lab Value can be used to
code this data item when no other information is available.
Note 2: Record the lab value of the AFP test results documented in the medical record after
orchiectomy but prior to adjuvant therapy. The lab value may be documented in a lab report, history
and physical, or clinical statement in the pathology report.
Note 3: If the initial post-orchiectomy AFP remains elevated, review subsequent tests and record the
lowest AFP value (normalization or plateau) prior to adjuvant therapy or before the value rises again.
Note 4: A lab value expressed in micrograms/liter (ug/L) is equivalent to the same value expressed in
ng/mL.
Note 5: If the lab value is expressed in IU/ml, use the following conversion: 1 ng/mL = 0.83 IU/mL.
o To calculate ng from IU/mL, divide the value for IU by 0.83.
o Example: 10 IU/mL: 10/0.83 = 12.04 ng/mL; 5 IU/mL: 5/0.83= 6.02 ng/mL
Note 6: If the pre-orchiectomy AFP was normal, a post-orchiectomy AFP may not be performed. In this
case, code XXXXX.9 should be recorded.
Note 7: If the only information available is a statement of elevated or normal, code XXXXX.9.

346 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 8: The same laboratory test should be used to record information in 3806: AFP Post-Orchiectomy
Range.
Code Description
0.0 0.0 nanograms/milliliter (ng/mL)
0.1-
99999.9
0.1 – 99,999.9 ng/mL
XXXXX.1 100,000 ng/mL or greater
XXXXX.7 Test ordered, results not in chart
XXXXX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XXXXX.8 may result
in an edit error.)
XXXXX.9 Not documented in medical record
No orchiectomy performed
AFP (Alpha Fetoprotein) Post-Orchiectomy Lab Value not assessed or unknown if
assessed
Return to Schema ID Table

347 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3806: AFP Post-Orchiectomy Range
Item Length: 1
NAACCR Item #: 3806
XML Parent-NAACCR ID: Tumor-afpPostOrchiectomyRange
NAACCR Alternate Name: AFP (Alpha Fetoprotein) Post-Orchiectomy Range
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
AFP (Alpha Fetoprotein) Post-Orchiectomy Range identifies the range category of the lowest AFP value
measured post-orchiectomy. AFP is a serum tumor marker that is often elevated in patients with
nonseminomatous germ cell tumors of the testis. The Post-Orchiectomy lab value is used to monitor
response to therapy.
Rationale
AFP (Alpha Fetoprotein) Post-Orchiectomy Range is a Registry Data Collection Variable in AJCC. AFP
(Alpha Fetoprotein) Post-Orchiectomy Range is used to assign the S Category Pathological and was
previously collected as Testis CS SSF #13.
See Alpha-fetoprotein (AFP) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the AFP (Alpha Fetoprotein) Post-Orchiectomy Range can be used to
code this data item when there is no other information available.
Note 2: Record the range of the AFP test as documented in the medical record after orchiectomy but
prior to adjuvant therapy. The lab value may be documented in a lab report, history and physical, or
clinical statement in the pathology report.
Note 3: If the initial post-orchiectomy AFP remains elevated, review subsequent tests and record the
lowest AFP value (normalization or plateau) prior to adjuvant therapy or before the value rises again.
Note 4: A lab value expressed in micrograms/liter (ug/L) is equivalent to the same value expressed in
nanograms/milliliter (ng/mL).
Note 5: If the lab value is expressed in IU/ml, use the following conversion: 1 ng/mL = 0.83 IU/mL.
o To calculate ng from IU/mL, divide the value for IU by 0.83.
o Example: 10 IU/mL: 10/0.83 = 12.04 ng/mL; 5 IU/mL: 5/0.83= 6.02 ng/mL
Note 6: If the pre-orchiectomy AFP was normal, a post-orchiectomy AFP may not be performed. In this
case, code 5 should be recorded.

348 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 7: The same laboratory test should be used to record information in 3805: AFP Post-Orchiectomy
Lab Value.
Code Description
0 Within normal limits
1 Above normal and less than 1,000 nanograms/milliliter (ng/mL)
2 1,000 -10,000 ng/mL
3 Greater than 10,000 ng/mL
4 Post-Orchiectomy alpha fetoprotein (AFP) stated to be elevated
5 Post-Orchiectomy alpha fetoprotein (AFP) unknown or not done but pre-orchiectomy AFP was
normal
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
No orchiectomy performed
AFP (Alpha Fetoprotein) Post-Orchiectomy Range not assessed or unknown if assessed
Return to Schema ID Table

349 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
Human Chorionic Gonadotropin (hCG) (Testis)
Definition
Human chorionic gonadotropin (hCG) is a hormone produced by the placenta and some germ cell
tumors. Two subunits, alpha and beta, can be measured in blood or serum. The alpha subunit is a non-
specific marker for pancreatic and pituitary tumors. Beta-hCG levels are never found in normal healthy
men. When the presence of beta-hCG is detected in serum, it always indicates a malignancy. Beta-hCG is
secreted by some non- seminomatous germ cell tumors and mixed tumors and is used with AFP to
identify the specific cell type of testicular cancer. Beta-hCG is also useful in monitoring response to
therapy. After orchiectomy, the hCG should be undetectable within 5 to 8 days. If elevated hCG persists,
this is an indication of residual tumor.
For Testis, there are 4 data items that record information on hCG.
3846: hCG Post-Orchiectomy Lab Value
3847: hCG Post-Orchiectomy Range
3848: hCG Pre-Orchiectomy Lab Value
3849: hCG Pre-Orchiectomy Range
Coding Guidelines
hCG Pre-Orchiectomy Lab Value and Range
Assign the code for the highest hCG value and corresponding hCG range prior to orchiectomy. In
the event an orchiectomy is not performed, or systemic treatment precedes an orchiectomy,
code the highest hCG value and corresponding range prior to any systemic treatment. The hCG
Range is a category used to group the lab values into 3 ranges: 1, 2, and 3. The pre-orchiectomy
hCG lab value and the pre-orchiectomy hCG range should be from the same test. If the clinician
states an S value rather than a lab value, code unknown (code XXXX.9) for the hCG pre-
orchiectomy lab value and unknown (code 9) for the hCG pre-orchiectomy range.
hCG Post-Orchiectomy Lab Value and Range
Assign the code for the lowest hCG value and corresponding hCG range after orchiectomy but
prior to adjuvant treatment. The half-life of human chorionic gonadotropic is 1 to 3 days, but it
may take much longer for this tumor marker to return to normal. If the first post-orchiectomy
hCG remains elevated, continue reviewing subsequent lab work and record the lowest hCG
value (normalization or plateau) prior to adjuvant treatment or before the value rises again. The
hCG Range is a category used to group the lab values into 3 ranges: 1, 2, and 3. The post-
orchiectomy hCG lab value and the post-orchiectomy hCG range should be from the same test.
If the clinician states an S value rather than a lab value, code unknown (code XXXX.9) for the
hCG post-orchiectomy lab value and unknown (code 9) for the hCG post-orchiectomy range.

350 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Categories used for Pre- and Post-Orchiectomy hCG Range are:
Code Description
0 Within normal limits
1 Above normal and less than 5,000 milli-International Units/milliliter (mIU/mL)
2 5,000 - 50,000 mIU/mL
3 Greater than 50,000 mIU/mL
Examples for hCG Pre-Orchiectomy and hCG Post-Orchiectomy Lab Value and Range
For these examples, the lab’s normal reference range for hCG = 0-5 mIU/mL
Examples Lab Value Code Range Code
2.0 mIU/mL 2.0 0
412 mIU/mL 412.0 1
6213 mIU/mL 6213.0 2
14,724 mIU/mL
14724.0
3
108,325 mIU/mL
XXXXX.1
3
Physician states “hCG elevated,” but no value documented XXXXX.9 4
S value stated (no other information available) XXXXX.9 9
No AFP test done, or unknown if done XXXXX.9 9
Additional Information
Source documents: clinical laboratory report (blood or serum test), sometimes in history and
physical or clinical statement in pathology report
For further information, refer to the Testis cancer protocol published by the College of American
Pathologists for the AJCC Staging System Testis
Other names: Human chorionic gonadotropin, b-hCG, beta subunit HCG, beta hCG, -hCG
Normal Reference Range
o < 2 ng/ml (SI: < 2 µg/L or < 2 ug/L) 1 ng/ml of HCG is approximately 5 mIU/ml.
o < 5 mIU/mL (< 5 IU/L) To record mIU/mL in ng/ml, divide the test result by 5.
Measurements: International Units/liter (IU/L) is equivalent to milli-International Units per
milliliter (mIU/ml)
Return to Schema ID Table

351 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3848: hCG Pre-Orchiectomy Lab Value
Item Length: 7
NAACCR Item #: 3848
XML Parent-NAACCR ID: Tumor-hcgPreOrchiectomyLabValue
NAACCR Alternate Name: hCG (Human Chorionic Gonadotropin) Pre-Orchiectomy Lab Value
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
hCG (Human Chorionic Gonadotropin) Pre-Orchiectomy Lab Value refers to the hCG value measured
prior to treatment. hCG is a serum tumor marker that is often elevated in patients with
nonseminomatous germ cell tumors of the testis.
Rationale
hCG (Human Chorionic Gonadotropin) Pre-Orchiectomy Lab Value is a Registry Data Collection Variable
in AJCC. It was previously collected as Testis CS SSF #8.
See Human Chorionic Gonadotropin (hCG) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the hCG (Human Chorionic Gonadotropin) Pre-Orchiectomy Lab Value
can be used to code this data item when no other information is available.
Note 2: Record the lab value of the highest hCG test result documented in the medical record prior to
orchiectomy or prior to any systemic treatment. The lab value may be documented in a lab report,
history and physical, or clinical statement in the pathology report.
Note 3: A lab value expressed in International Units/liter (IU/L) is equivalent to the same value
expressed in milli-International Units/milliliter (mIU/mL).
Note 4: The same laboratory test should be used to record information in 3849: hCG Pre-Orchiectomy
Range.
Code Description
0.0 0.0 milli-International Units/milliliter (mIU/mL)
0.1-99999.9 0.1 – 99,999.9 mIU/mL
XXXXX.1 100,000 mIU/mL or greater
XXXXX.7 Test ordered, results not in chart
XXXXX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XXXXX.8 may result
in an edit error.)
XXXXX.9 Not documented in medical record
hCG (Human Chorionic Gonadotropin) Pre-orchiectomy Lab Value not assessed or
unknown if assessed
Return to Schema ID Table

352 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3849: hCG Pre-Orchiectomy Range
Item Length: 1
NAACCR Item #: 3849
XML Parent-NAACCR ID: Tumor-hcgPreOrchiectomyRange
NAACCR Alternate Name: hCG
Schema(s):
00590: Testis (2018+)
Description
Human Chorionic Gonadotropin (hCG) Pre-Orchiectomy Range identifies the range category of the
highest hCG value measured prior to treatment. hCG is a serum tumor marker that is often elevated in
patients with nonseminomatous germ cell tumors of the testis.
Rationale
hCG (Human Chorionic Gonadotropin) is a Registry Data Collection Variable in AJCC. hCG (Human
Chorionic Gonadotropin) Pre-Orchiectomy Range is used to assign the S Category Clinical and was
previously collected as Testis CS SSF #9.
See Human Chorionic Gonadotropin (hCG) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the hCG (Human Chorionic Gonadotropin) Pre-Orchiectomy Range can
be used to code this data item when no other information is available.
Note 2: Record the range of the highest hCG test result documented in the medical record prior to
orchiectomy or prior to any systemic treatment. The lab value may be documented in a lab report,
history and physical, or clinical statement in the pathology report.
Note 3: A lab value expressed in International Units/liter (IU/L) is equivalent to the same value
expressed in milli-International Units/milliliter (mIU/mL).
Note 4: The same laboratory test should be used to record information in 3848: hCG Pre-Orchiectomy
Lab Value.
Code Description
0 Within normal limits
1 Above normal and less than 5,000 milli-International Units/milliliter (mIU/mL)
2 5,000 - 50,000 mIU/mL
3 Greater than 50,000 mIU/mL
4 Pre-orchiectomy human chorionic gonadotropin (hCG) stated to be elevated
7 Test ordered, results not in chart
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9
Not documented in medical record
hCG pre-orchiectomy range not assessed or unknown if assessed
Return to Schema ID Table

353 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3846: hCG Post-Orchiectomy Lab Value
Item Length: 7
NAACCR Item #: 3846
XML Parent-NAACCR ID: Tumor-hcgPostOrchiectomyLabValue
NAACCR Alternate Name: hCG (Human Chorionic Gonadotropin) Post-Orchiectomy Lab Value
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
hCG (Human Chorionic Gonadotropin) Post-Orchiectomy Lab Value refers to the lowest hCG value
measured post-orchiectomy. hCG is a serum tumor marker that is often elevated in patients with
nonseminomatous germ cell tumors of the testis. The Post-Orchiectomy lab value is used to monitor
response to therapy.
Rationale
hCG (Human Chorionic Gonadotropin) Post-Orchiectomy Lab Value is a Registry Data Collection Variable
in AJCC. It was previously collected as Testis CS SSF #14.
See Human Chorionic Gonadotropin (hCG) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the hCG (Human Chorionic Gonadotropin) Post-Orchiectomy Lab Value
can be used to code this data item when no other information is available.
Note 2: Record the value of the hCG test as documented in the medical record after orchiectomy but
prior to adjuvant therapy. The lab value may be documented in a lab report, history and physical, or
clinical statement in the pathology report.
Note 3: If the initial post-orchiectomy hCG remains elevated, review subsequent tests and record the
lowest hCG value (normalization or plateau) prior to adjuvant therapy or before the value rises again.
Note 4: A lab value expressed in International Units/liter (IU/L) is equivalent to the same value
expressed in milli-International Units/milliliter (mIU/mL).
Note 5: If the pre-orchiectomy hCG was normal, a post-orchiectomy hCG may not be performed. In this
case, code XXXXX.9 should be recorded.
Note 6: If the only information available is a statement of elevated or normal, code XXXXX.9.
Note 7: The same laboratory test should be used to record information in 3847: hCG Post-Orchiectomy
Range.

354 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0.0 0.0 milli-International Units/milliliter (mIU/mL)
0.1
-
99999.9
0.1
–
99,999.9 mIU/mL
XXXXX.1 100,000 mIU/mL or greater
XXXXX.7 Test ordered, results not in chart
XXXXX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XXXXX.8 may result
in an edit error.)
XXXXX.9 Not documented in medical record
No orchiectomy performed
hCG (Human Chorionic Gonadotropin) Post-orchiectomy Lab Value not assessed or
unknown if assessed
Return to Schema ID Table

355 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3847: hCG Post-Orchiectomy Range
Item Length: 1
NAACCR Item #: 3847
XML Parent-NAACCR ID: Tumor-hcgPostOrchiectomyRange
NAACCR Alternate Name: hCG (Human Chorionic Gonadotropin) Post-Orchiectomy Range
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
Human Chorionic Gonadotropin (hCG) Post-Orchiectomy Range identifies the range category of the
lowest hCG value measured post-orchiectomy. hCG is a serum tumor marker that is often elevated in
patients with nonseminomatous germ cell tumors of the testis. The Post-Orchiectomy lab value is used
to monitor response to therapy.
Rationale
hCG (Human Chorionic Gonadotropin) is a Registry Data Collection Variable in AJCC. hCG (Human
Chorionic Gonadotropin) Post-orchiectomy Range is used to assign the S Category Pathological and was
previously collected as Testis CS SSF #15.
See Human Chorionic Gonadotropin (hCG) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the hCG (Human Chorionic Gonadotropin) Post-orchiectomy Range can
be used to code this data item when there is no other information available.
Note 2: Record the range of the hCG test as documented in the medical record after orchiectomy but
prior to adjuvant therapy. The lab value may be documented in a lab report, history and physical, or
clinical statement in the pathology report.
Note 3: If the initial post-orchiectomy hCG remains elevated, review subsequent tests and record the
lowest hCG value (normalization or plateau) prior to adjuvant therapy or before the value rises again.
Note 4: A lab value expressed in International Units/liter (IU/L) is equivalent to the same value
expressed in milli-International Units/milliliter (mIU/mL).
Note 5: If the pre-orchiectomy hCG was normal, a post-orchiectomy hCG may not be performed. In this
case, code 5 should be recorded.
Note 6: The same laboratory test should be used to record information in 3846: hCG Post-Orchiectomy
Lab Value.

356 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 Within normal limits
1 Above normal and less than 5,000 milli-International Units/milliliter (mIU/mL)
2 5,000 - 50,000 mIU/mL
3 Greater than 50,000 mIU/mL
4 Post-orchiectomy human chorionic gonadotropin (hCG) stated to be elevated
5
Post
-
Orchiectomy human chorionic gonadotropin (hCG) unknown or not done but pre
-
orchiectomy hCG was normal
7
Test ordered, results not in chart
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
No orchiectomy performed
hCG (Human Chorionic Gonadotropin) Post-orchiectomy Range not assessed or unknown if
assessed
Return to Schema ID Table

357 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
Lactate Dehydrogenase (LDH) (Testis)
Definition
When cells (normal or tumor) are damaged or destroyed, an enzyme called lactate dehydrogenase (LDH)
is released into the bloodstream. LDH is an indirect indication of possible tumor burden or damage to
an organ, which may be caused by metastatic involvement of liver or lung, or a myocardial
infarction. The total LDH should be the test value that is coded, but there are five fractions of LDH that
measure tissue specific cellular damage: LD1 and LD2: heart, red blood cells and kidneys; LD3: lung; LD4
and LD5: liver, skin and skeletal muscles. LDH is elevated in 60% of patients with non-seminomatous
germ cell tumors of the testis. LDH is not a screening test, nor is it diagnostic of melanoma, ocular
adnexal lymphoma, or testicular cancer.
For testis, only the LDH Range is coded. LDH is non-specific for testicular cancer. Although part of the
criteria for the S category in the TNM system, LDH is not routinely performed unless the patient has
evidence of bulky or distant disease.
For Testis, there are 2 data items that record information on LDH.
3867: LDH Post-Orchiectomy Range
3868: LDH Pre-Orchiectomy Range
Coding guidelines
LDH Pre-Orchiectomy Range
The LDH Range is a category used to group the lab values into 3 ranges: 1, 2, and 3. Code the range of
the highest LDH value prior to orchiectomy, based on the reference range used by the lab. In the event
an orchiectomy is not performed, or systemic treatment precedes an orchiectomy, code the range of
the highest LDH value prior to any systemic treatment. If the clinician states an S value rather than a lab
value, code unknown (code 9).
LDH Post-Orchiectomy Range
Code the range of the lowest LDH after orchiectomy but prior to adjuvant treatment. If the first post-
orchiectomy LDH remains elevated, continue reviewing subsequent lab work and record the lowest LDH
value (normalization or plateau) prior to adjuvant treatment or before the value rises again. If the
clinician states an S value rather than a lab value, code unknown (code 9).
Categories used for Pre- and Post-Orchiectomy LDH Range are:
Code Description
0 Within normal limits
1
Less than 1.5 x N
(Less than 1.5 times the upper limit of normal for LDH)
2
1.5 to 10 x N
(Between 1.5 and 10 times the upper limit of normal for LDH)

358 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
3 Greater than 10 x N
(Greater than 10 times the upper limit of normal for LDH)
To calculate whether the lab result is in a particular range, multiply the lab’s upper limit of normal
(usually stated on the report) times the stated multiplier. If the test is elevated, determine whether it is
less than 1.5 times the upper limit of normal (code 1), between 1.5 and 10 times the upper limit of
normal (code 2), or more than 10 times the upper limit of normal (code 3).
Examples for LDH Pre-Orchiectomy and Post-Orchiectomy Range
For these examples, the lab’s normal reference range for LDH = 100-225
o 1.5 X 225 (upper limit of normal) = 337.5
o 10 x 225 (upper limit of normal) = 2250
Therefore, for this lab, a value that is elevated and up to 337 = code 1, a value from 338 to 2250 = code
2, and a value greater than 2250 = code 3.
Examples Code
118
(within normal range 100-225)
0
282
(elevated but less than 337.5)
1
1081
(elevated and between 337.5 and 2250)
2
2795
(elevated and greater than 2250)
3
Physician states “LDH elevated,” but no value documented 4
No LDH test done, or unknown if done 9
S value stated (no other information available) 9
Additional Information
Source documents: clinical laboratory report; may be included in a liver or hepatic panel/profile,
a cardiac panel, or a general metabolic panel of tests
For further information, refer to the Testis cancer protocol published by the College of American
Pathologists for the AJCC Staging System Testis
Other names: LD, Lactate dehydrogenase, lactase dehydrogenase, lactic acid dehydrogenase
Normal reference range: varies widely by laboratory, patient age, and the units of
measurement.
Return to Schema ID Table

359 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3868: LDH Pre-Orchiectomy Range
Item Length: 1
NAACCR Item #: 3868
XML Parent-NAACCR ID: Tumor-ldhPreOrchiectomyRange
NAACCR Alternate Name: LDH (Lactate Dehydrogenase) Pre-Orchiectomy Range
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
Lactate Dehydrogenase (LDH) Range identifies the range category of the highest LDH value measured
prior to treatment. LDH is a nonspecific marker for testicular cancer that is elevated in some germ cell
tumors. This data item refers to the Pre-Orchiectomy range.
Rationale
LDH (Lactate Dehydrogenase) is a Registry Data Collection Variable in AJCC. LDH (Lactate
Dehydrogenase) Pre-Orchiectomy Range is used to assign the S Category Clinical and was previously
collected as Testis CS SSF #10.
See Lactate Dehydrogenase (LDH) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the LDH (Lactate Dehydrogenase) Pre-Orchiectomy Range can be used to
code this data item when no other information is available.
Note 2: Record the range of the highest LDH test result documented in the medical record prior to
orchiectomy or prior to any systemic treatment. The lab value may be documented in a lab report,
history and physical, or clinical statement in the pathology report.
Note 3: Of the three tumor markers, lactate dehydrogenase (LDH) is the least specific for testicular
cancer. The magnitude of LDH elevation directly correlates with Testis tumor burden.
Code Description
0 Within normal limits
1
Less than 1.5 x N
(Less than 1.5 times the upper limit of normal for LDH)
2
1.5 to 10 x N
(Between 1.5 and 10 times the upper limit of normal for LDH)
3
Greater than 10 x N
(Greater than 10 times the upper limit of normal for LDH)
4 Pre-Orchiectomy lactate dehydrogenase (LDH) range stated to be elevated
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit error.)
9 Not documented in medical record
LDH (Lactate Dehydrogenase) Pre-Orchiectomy Range not assessed or unknown if assessed
Return to Schema ID Table

360 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3867: LDH Post-Orchiectomy Range
Item Length: 1
NAACCR Item #: 3867
XML Parent-NAACCR ID: Tumor-ldhPostOrchiectomyRange
NAACCR Alternate Name: LDH (Lactate Dehydrogenase) Post-Orchiectomy Range
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
LDH (Lactate Dehydrogenase) Post-Orchiectomy Range identifies the range category of the lowest LDH
value measured post-orchiectomy. LDH is a nonspecific marker for testicular cancer that is elevated in
some germ cell tumors. The Post-Orchiectomy lab value is used to monitor response to therapy.
Rationale
LDH (Lactate Dehydrogenase) is a Registry Data Collection Variable in AJCC. LDH (Lactate
Dehydrogenase) Post-Orchiectomy Range is used to assign the S Category Pathological and was
previously collected as Testis CS SSF #16.
See Lactate Dehydrogenase (LDH) (Testis) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of the LDH (Lactate Dehydrogenase) Post-Orchiectomy Range can be used
to code this data item when there is no other information available.
Note 2: Record the range of the LDH test as documented in the medical record after orchiectomy but
prior to adjuvant therapy. The lab value may be documented in a lab report, history and physical, or
clinical statement in the pathology report.
Note 3: If the initial post-orchiectomy LDH remains elevated, review subsequent tests and record the
lowest LDH value (normalization or plateau) prior to adjuvant therapy or before the value rises again.
Note 4: Of the three tumor markers, lactate dehydrogenase (LDH) is the least specific for testicular
cancer. The magnitude of LDH elevation directly correlates with Testis tumor burden.
Note 5: If the pre-orchiectomy LDH was normal, a post-orchiectomy LDH may not be performed. In this
case, code 5 should be recorded.
Code Description
0 Within normal limits
1
Less than 1.5 x N
(Less than 1.5 times the upper limit of normal for LDH)
2
1.5 to 10 x N
(Between 1.5 and 10 times the upper limit of normal for LDH)

361 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
3 Greater than 10 x N
(Greater than 10 times the upper limit of normal for LDH)
4 Post-Orchiectomy lactate dehydrogenase (LDH) range stated to be elevated
5
Post
-
Orchiectomy lactage
dehydrogenase (LDH) unknown or not done but pre
-
orchiectomy
LDH was normal
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
No orchiectomy performed
LDH (Lactate Dehydrogenase) Post-Orchiectomy Range not assessed or unknown if assessed
Return to Schema ID Table

362 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3923: S Category Clinical
Item Length: 1
NAACCR Item #: 3923
XML Parent-NAACCR ID: Tumor-sCategoryClinical
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
S Category Clinical combines the results of pre-orchiectomy Alpha Fetoprotein (AFP), Human Chorionic
Gonadotropin (hCG) and Lactate Dehydrogenase (LDH) into a summary S value.
Rationale
S Category Clinical is required for prognostic stage grouping in Chapter 59 Testis. It is a new data item for
cases diagnosed 1/1/2018+.
Additional Information
For further information, refer to the Testis cancer protocol published by the College of American
Pathologists for the AJCC Staging System Testis
Coding Instructions and Codes
Note 1: Code the S category as described by the physician. If the S category determined by available lab
values or calculated by vendor software differs from the physician statement of the S category, the
physician’s statement takes precedence.
Note 2: Code the pre-orchiectomy S category according to the table below. This table is also available in
AJCC 8
th
edition, Chapter 59, Testis.
For AFP, a lab value expressed in micrograms per liter (ug/L) is equivalent to the same value
expressed in nanograms per milliliter (ng/ml).
Note 3: Clinical stage values are those based on physician statement or lab values at diagnosis, prior to
orchiectomy, and prior to any systemic treatment.
Note 4: All three lab values are needed for S0-S1. Only one elevated test is needed to assign S2-3. If any
individual test is not available and none of the available tests results meets the S2-3 criterion for that
test, assign code 9 (SX).

363 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 S0: Marker study levels within normal levels
1 S1: At least one of these values is elevated AND
LDH less than 1.5 x N* AND
hCG (mIU/L) less than 5,000 AND
AFP (ng/mL) less than 1,000
2 S2:
LDH 1.5 x N* to 10 x N* OR
hCG (mIU/L) 5,000 to 50,000 OR
AFP (ng/mL) 1,000 to 10,000
3 S3: Only one elevated test is needed
LDH greater than 10 x N* OR
hCG (mIU/mL) greater than 50,000 OR
AFP (ng/mL) greater than 10,000
9
SX:
Not documented in medical record
S Category Clinical not assessed or unknown if assessed
*N indicates the upper limit of normal for the LDH assay.
Return to Schema ID Table

364 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00590: Testis (2018+)
3924: S Category Pathological
Item Length: 1
NAACCR Item #: 3924
XML Parent-NAACCR ID: Tumor-sCategoryPathological
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00590: Testis (2018+)
Description
S Category Pathological combines the results of post-orchiectomy Alpha Fetoprotein (AFP), Human
Chorionic Gonadotropin (hCG) and Lactate Dehydrogenase (LDH) into a summary S value.
Rationale
S Category Pathological is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 59 Testis. It
is a new data item for cases diagnosed 1/1/2018+.
Additional Information
For further information, refer to the Testis cancer protocol published by the College of American
Pathologists for the AJCC Staging System Testis
Coding Instructions and Codes
Note 1: Code the S category as described by the physician. If the S category determined by available lab
values or calculated by vendor software differs from the physician statement of the S category, the
physician’s statement takes precedence.
Note 2: Code the post-orchiectomy S category according to the table below. This table is also available
in AJCC 8
th
edition, Chapter 59, Testis.
For AFP, a lab value expressed in micrograms per liter (ug/L) is equivalent to the same value
expressed in nanograms per milliliter (ng/ml).
Note 3: Pathological stage values are those based on physician statement or lab values after
orchiectomy and prior to adjuvant therapy.
Note 4: If the initial post-orchiectomy lab values remain elevated, review the subsequent tests and use
the lowest lab values (normalization or plateau) prior to adjuvant therapy or before the value rises
again.
Note 5: All three lab values are needed for S0-S1. Only one elevated test is needed to assign S2-3. If any
individual test is not available and none of the available tests results meets the S2-3 criterion for that
test, assign code 9 (SX).
Note 6: When all the serum tumor markers are normal pre-orchiectomy and they are not repeated post-
orchiectomy, code 5.

365 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0 S0: Marker study levels within normal levels
1 S1: At least one of these values is elevated AND
LDH less than 1.5 x N* AND
hCG (mIU/L) less than 5,000 AND
AFP (ng/mL) less than 1,000
2 S2
LDH 1.5 x N* to 10 x N* OR
hCG (mIU/L) 5,000 to 50,000 OR
AFP (ng/mL) 1,000 to 10,000
3 S3: Only one elevated test is needed
LDH greater than 10 x N* OR
hcG (mIU/mL) greater than 50,000 OR
AFP (ng/mL) greater than 10,000
5
Post
-
orchiectomy serum tumor markers
unknown or not done but pre
-
orchiectomy
serum tumor markers were normal
9
SX:
Not documented in medical record
S Category Pathological not assessed or unknown if assessed
*N indicates the upper limit of normal for the LDH assay.
Return to Schema ID Table
366 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
URINARY TRACT

367 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00600: Kidney Parenchyma (2018+)
3864: Invasion Beyond Capsule
Item Length: 1
NAACCR Item #: 3864
XML Parent-NAACCR ID: Tumor-invasionBeyondCapsule
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00600: Kidney (2018+)
Description
Kidney Tumor Extension pertains to the pathologically confirmed invasion of the tumor beyond the
fibrous capsule in which the kidney is enclosed.
Rationale
Kidney Tumor Extension into specific tissues for Kidney is a Registry Data Collection Variable in AJCC. It
was previously collected as Kidney, CS SSF #1.
Definition
This data item collects additional information on the description of tumor spread (invasion beyond
capsule) as documented in the pathology report. Do not include clinical findings in this field.
Coding guidelines
Code 0: There is no invasion beyond capsule
o If surgical resection is done and tumor is “confined to kidney” and staging is based on
size, then there has been no invasion through the capsule (no invasion into perinephric
fat)
Code 1: Perinephric fat, which is the layer of fat (adipose tissue) outside the renal capsule but
inside Gerota’s fascia
Code 2: Renal sinus, which is the elongated oval indentation in the renal parenchyma occupied
by the renal pelvis, renal calyces, blood vessels, nerves, and perisinus fat
o Synonyms include: renal hilum, renal sinus fat, medial invasion
Code 3: Gerota’s fascia (Gerota’s capsule), which is a fibrous envelope of tissue that surrounds
the kidney
Code 4: Any combination of codes 1-3
Code 5: Invasion beyond the capsule, NOS
Code 9 when
o There is no documentation in the medical record
o Clinical diagnosis only
o Evaluation of capsule invasion not done or unknown if done

368 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
For further information, refer to the Kidney cancer protocol published by the College of
American Pathologists for the AJCC Staging System Kidney
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
is no mention of invasion beyond capsule, the registrar could assume that it was negative and
code appropriately. For the SSDI, this assumption cannot be made. There must be a statement
that invasion beyond capsule is not present to code 0.
Coding Instructions and Codes
Note 1: Physician statement of pathologically confirmed invasion of the tumor beyond the fibrous
capsule (invasion beyond capsule) can be used to code this data item.
Note 2: Information about invasion beyond the capsule is collected in primary tumor as an element in
anatomic staging. It is also collected in this field as it may have an independent effect on prognosis.
If surgical resection is done and tumor is “confined to kidney” and staging is based on size, then
there has been no invasion through the capsule (no invasion into perinephric fat)
Note 3: Perinephric/sinus fat invasion should be confirmed microscopically and is invasion into fat by
tumor cells, with or without desmoplastic reaction, and vascular invasion into perinephric/sinus soft
tissue.
Synonyms include: renal hilum, renal sinus fat, medial invasion
Note 4: Record invasion beyond capsule as documented in the pathology report.
Note 5: Do not use imaging findings to code this data item.
Note 6: Code 9 if surgical resection of the primary site is performed and there is no mention of invasion
beyond capsule.
Code Description
0 Invasion beyond capsule not identified
1 Perinephric (beyond renal capsule) fat or tissue
2 Renal sinus
3 Gerota’s fascia
4 Any combination of codes 1-3
5 Invasion beyond capsule, NOS
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Invasion beyond capsule not assessed or unknown if assessed
No surgical resection of primary site is performed
Return to Schema ID Table

369 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00600: Kidney Parenchyma (2018+)
3886: Major Vein Involvement
Item Length: 1
NAACCR Item #: 3886
XML Parent-NAACCR ID: Tumor-majorVeinInvolvement
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00600: Kidney
Description
Major vein involvement pertains to the invasion of the kidney tumor into major veins.
Rationale
Involvement of major veins for Kidney is a Registry Data Collection Variable in AJCC. It was previously
collected as Kidney, CS SSF #2.
Definition
Involvement of veins from a renal cancer has prognostic implications because tumor cells can more
easily disseminate through the bloodstream. This data item records information about the presence and
level of involvement of specific major blood vessels. Do not code microscopically identified involvement
of small unnamed blood vessels within the kidney; this information is coded in the field Lymph-Vascular
Invasion (LVI). The tumor may be described as a thrombus, a cluster of tumor cells presents in the center
of the vein but not attached to the wall of the vein. Tumor spread may resemble mud extruding along
the inside of a pipe. Direct tumor invasion of the wall of the inferior vena cava is not coded in this field.
Record the code that best describes involvement of the renal vein and/or inferior vena cava (IVC) as
described in the pathology report. Do not include clinical findings in this field.
Coding guidelines
Code 0: There is no involvement of the major veins
o If surgical resection is done and tumor is “confined to kidney” and staging is based on
size, then there is no involvement of major veins
Code 1: Involvement of the renal vein or segmental branches
Code 2: Involvement of the inferior vena cava (IVC)
Code 3: Involvement of major veins, but not specified which one (renal vein, segmental
branches or inferior vena cava (IVC))
Code 4: Involvement of more than one vein (any combination of codes 1-3)
Code 9 when
o There is no documentation in the medical record
o Clinical diagnosis only
o Evaluation of major vein involvement not done or unknown if done

370 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
For further information, refer to the Kidney cancer protocol published by the College of
American Pathologists for the AJCC Staging System Kidney
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of major vein involvement, the registrar could assume that it was negative and
code appropriately. For the SSDI, this assumption cannot be made. There must be a statement
that major vein involvement is not present to code 0.
Coding Instructions and Codes
Note 1: Physician statement of Major Vein Involvement can be used to code this data item. The major
veins include the renal vein or its segmental branches, and the inferior vena cava.
Note 2: Information about major vein involvement beyond the kidney is collected in primary tumor as
an element in anatomic staging. It is also collected in this field as it may have an independent effect on
prognosis.
If surgical resection is done and tumor is “confined to kidney” and staging is based on size, then
there is no involvement of major veins
Note 3: Record the involvement of specific named veins as documented in the pathology report. Do not
code invasion of small unnamed vein(s) of the type collected as lymph-vascular invasion. Lymph-vascular
invasion is usually only seen microscopically.
Note 4: Do not use imaging findings to code this data item.
Note 5: Code 9 if surgical resection of the primary site is performed and there is no mention of major
vein involvement.
Code
Description
0 Major vein involvement not present/not identified
1 Renal vein or its segmental branches
2 Inferior vena cava (IVC)
3 Major vein invasion, NOS
4 Any combination of codes 1-3
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Vein involvement not assessed or unknown if assessed
No surgical resection of primary site is performed
Return to Schema ID Table

371 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00600: Kidney Parenchyma (2018+)
3861: Ipsilateral Adrenal Gland Involvement
Item Length: 1
NAACCR Item #: 3861
XML Parent-NAACCR ID: Tumor-ipsilateralAdrenalGlandInvolve
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00600: Kidney (2018+)
Description
Ipsilateral adrenal gland involvement pertains to direct extension of the tumor into the ipsilateral
adrenal gland (continuous) or ipsilateral adrenal gland involvement by a separate nodule
(discontiguous).
Rationale
Ipsilateral adrenal gland involvement for Kidney is a Registry Data Collection Variable in AJCC. It was
previously collected as Kidney, CS SSF #3.
Definition
The adrenal gland is contained within Gerota’s fascia and is contiguous with the kidney, but it has its
own lymphatic and vascular drainage systems. Involvement of the ipsilateral (same side) adrenal gland
by kidney tumor—an adverse prognostic indicator—may be by direct extension (contiguous) or
hematogenous (through the bloodstream; discontiguous). Do not include clinical findings in this field.
Coding guidelines
Code 0: There is no involvement of the ipsilateral adrenal gland
o If surgical resection is done and tumor is “confined to kidney” and staging is based on
size, then there is no involvement of the adrenal gland
Code 1: Ipsilateral adrenal gland involved by direct extension (contiguous involvement)
Code 2: Ipsilateral adrenal gland involved by separate nodule (discontiguous involvement)
Code 3: Ipsilateral adrenal gland involvement by contiguous and discontiguous involvement
Code 4: Ipsilateral adrenal gland involvement, unknown if contiguous or discontiguous
involvement
Code 9 when
o There is no documentation in the medical record
o Clinical diagnosis only
o Evaluation of ipsilateral adrenal gland involvement not done or unknown if done
Additional Information
Source documents: pathology report
For further information, refer to the Kidney cancer protocol published by the College of
American Pathologists for the AJCC Staging System Kidney
Other names: suprarenal gland; same side (ipsilateral)

372 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of ipsilateral gland involvement, the registrar could assume that it was negative
and code appropriately. For the SSDI, this assumption cannot be made. There must be a
statement that ipsilateral gland involvement is not present to code 0.
Coding Instructions and Codes
Note 1: Physician statement of Ipsilateral Adrenal Gland Involvement can be used to code this data item.
Note 2: Information about contiguous ipsilateral adrenal gland involvement is collected in primary
tumor, and discontiguous ipsilateral adrenal gland involvement is collected in distant metastasis, as
elements in anatomic staging. This information is also collected in this field as it may have an
independent effect on prognosis.
If surgical resection is done and tumor is “confined to kidney” and staging is based on size, then
there is no involvement of the adrenal gland
Note 3: Record ipsilateral adrenal gland involvement as documented in the pathology report.
Note 4: Do not use imaging findings to code this data item.
Note 5: Code 9 if surgical resection of the primary site is performed and there is no mention of
ipsilateral adrenal gland involvement.
Code Description
0 Ipsilateral adrenal gland involvement not present/not identified
1 Adrenal gland involvement by direct involvement (contiguous involvement)
2 Adrenal gland involvement by separate nodule (discontiguous involvement)
3 Combination of code 1-2
4 Ipsilateral adrenal gland involvement, unknown if direct involvement or separate nodule
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Ipsilateral adrenal gland not resected
Ipsilateral adrenal gland involvement not assessed or unknown if assessed
No surgical resection of primary site is performed
Return to Schema ID Table

373 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00600: Kidney Parenchyma (2018+)
3925: Sarcomatoid Features
Item Length: 3
NAACCR Item #: 3925
XML Parent-NAACCR ID: Tumor-sarcomatoidFeatures
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00600: Kidney (2018+)
Description
Sarcomatoid features: present or absent and percentage refers to the observation of sheets and
fascicles of malignant spindle cells in a kidney tumor which can occur across all histologic subtypes. The
percentage of sarcomatoid component has been shown to correlate with cancer-specific mortality.
Rationale
Sarcomatoid features for Kidney is a Registry Data Collection Variable in AJCC. It was previously
collected as Kidney, CS SSF #4.
Definition
The presence of sarcomatoid or spindle cell features in a kidney tumor is a strong adverse prognostic
factor. There is a specific ICD-O morphology code for renal cell carcinoma, sarcomatoid or spindle cell
(8318/3), but this data item documents any sarcomatoid or spindle cell features in any renal cell cancer.
Note: This data item applies to carcinomas only; rare sarcomas of the kidney should not be
coded in this field
Code the percentage of sarcomatoid features documented anywhere in the pathology report
Coding guidelines
Record whether Sarcomatoid features are present or absent.
Code 000 when the pathology report states that there are no sarcomatoid features
Code 001-100 code exact percentage of sarcomatoid features appropriately [1% (001) to 100%
(100)]
Code R01-R05 when only range documented (specific percentage not available)
Code XX5 when the only information available about Sarcomatoid features is from a metastatic site
Code XX6 when sarcomatoid features present, percentage unknown
Code XX7 when histology is not renal cell carcinoma
Code XX9 when
o Not documented in medical record
o No surgical resection done
o Pathology report not available
o Sarcomatoid features not evaluated (not assessed)
o Unknown if Sarcomatoid Features evaluated (assessed)

374 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
For further information, refer to the Kidney cancer protocol published by the College of
American Pathologists for the AJCC Staging System Kidney
Other names: spindle cell features
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of sarcomatoid features, the registrar could assume that they were not present
and code appropriately. For the SSDI, this assumption cannot be made. There must be a
statement that sarcomatoid features are not present to code 000.
Coding Instructions and Codes
Note 1: Physician statement of Sarcomatoid Features can be used to code this data item.
Note 2: Sarcomatoid morphology may be manifested by any renal cell carcinoma. The presence of
sarcomatoid component in a renal cell carcinoma may be prognostically important.
Note 3: Sarcomatoid features is mostly seen with renal cell carcinoma (all variants); however, if it’s seen
with other histologies, it can be coded.
Note 4: Record the presence or absence of sarcomatoid features as documented anywhere in the
pathology report.
Note 5: Code XX5 when the only information available about Sarcomatoid features is from a metastatic
site.
Note 6: Do not use imaging findings to code this data item.
Note 7: Code XX9 if surgical resection of the primary site is performed and there is no mention of
sarcomatoid features.
Code Description
000 Sarcomatoid features not present/not identified
001-100 Sarcomatoid features 1-100%
R01 Sarcomatoid features stated as less than 10%
R02 Sarcomatoid features stated as range 10%-30% present
R03 Sarcomatoid features stated as a range 31% to 50% present
R04
Sarcomatoid features stated as a range 51% to 80% present
R05
Sarcomatoid features stated as greater than 80%
XX5 Sarcomatoid features present from metastatic site only AND
Sarcomatoid features not present, or unknown if present, in primary site
XX6 Sarcomatoid features present, percentage unknown
XX7 Not applicable: Not a renal cell carcinoma morphology
XX8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XX8 may result in an
edit error.)
XX9
Not documented in medical record
Sarcomatoid features not assessed or unknown if assessed
No surgical resection of primary site is performed
Return to Schema ID Table

375 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00631: Urethra (2018+)
3926: Schema Discriminator 1: Urethra/Prostatic Urethra
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00631: Urethra (2018+)
00633: Urethra-Prostatic (2018+)
Definition
Urethra (male and female) and prostatic urethra have the same ICD-O topography code (C680).
However, for purposes of stage grouping AJCC 8
th
edition, they each have different definitions for T or
primary tumor extension. A schema discriminator is necessary to distinguish between these primary
sites so that the appropriate sub(chapter)/schema is used.
Coding Instructions and Codes
Note: A schema discriminator is used to discriminate between urethra (male and female) and prostatic
urethra. Code the site in which the tumor arose.
00631: Urethra (see code 1)
o Subsites include: Urethra, NOS; Urethral gland, Cowper gland
00633: Urethra-Prostatic Urethra (see code 2)
o Subsites include: Prostatic urethra, Prostatic utricle
Code Description Schema ID#/Description
1 Male penile urethra
Female urethra
Urethral gland
Cowper gland
Urethra, NOS
00631: Urethra
2 Males only
Prostatic urethra
Prostatic utricle
00633: Urethra-Prostatic
Return to Schema ID Table
377 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
OPHTHALMIC SITES

378 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00640: Skin Eyelid (2018+)
3909: Perineural Invasion
Item Length: 1
NAACCR Item #: 3909
XML Parent-NAACCR ID: Tumor-perineuralInvasion
NAACCR Alternate Name: None
Active years: 2018+
AJCC 8th Edition Chapter(s):
Chapter 15: Cutaneous Carcinoma of the Head and Neck (2018+)
Chapter 20: Colon and Rectum (2018+)
Chapter 64: Eyelid Carcinoma (2018+)
Chapter 69: Lacrimal Gland (2018+)
Description
Perineural Invasion, within or adjacent to the primary tumor, is a negative prognostic factor for
cutaneous squamous cell carcinomas of the head and neck and carcinomas of the colon and rectum,
eyelid and lacrimal gland.
Rationale
Perineural Invasion is a Registry Data Collection Variable in AJCC. It was previously collected as Colon
and Rectum CS SSF #8 and Lacrimal Gland CS SSF #4.
Definition
Perineural invasion is infiltration of nerves in the area of the lesion by tumor cells or spread of tumor
along the nerve pathway. The presence of perineural invasion has been shown in several studies to be
an indicator of poor patient prognosis. Where positive findings like perineural invasion are expected to
be included in pathology reports, negative results can be assumed if they are not specifically addressed.
Code whether perineural invasion is present based on the description in the pathology report.
Additional Information
Source documents: pathology report
Other names: PNI, neurotropism
Change from Collaborative Stage v2 (CSv2): In CSv2, if pathology report was available and there
was no mention of perineural invasion, the registrar could assume that it was negative and code
appropriately. Per the SSDI as of 2018, this assumption cannot be made. There must be a
statement that perineural invasion is not present/negative to assign “negative.” (Code 0)
Coding Instructions and Codes
Note 1: Physician statement of microscopically confirmed perineural invasion can be used to code this
data item when no other information is available.

379 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 2: Code the presence or absence of perineural invasion by the primary tumor as documented in
the pathology report.
Note 3: Information on presence of perineural invasion can be taken from either a biopsy or resection.
Absence of perineural invasion can only be taken from a surgical resection pathology report.
Note 4: Code 9 if surgical resection of the primary site is performed and there is no mention of
perineural invasion.
Code Description
0
Perineural invasion not identified/not present
Non-invasive neoplasm (behavior /2)
1 Perineural invasion identified/present
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Pathology report does not mention perineural invasion
Cannot be determined by the pathologist
Perineural invasion not assessed or unknown if assessed
Return to Schema ID Table

381 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3926: Schema Discriminator 1: Melanoma Ciliary Body/Melanoma Iris
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Definition
Iris and ciliary body have the same ICD-O topography code (C694). However, for purposes of stage
grouping AJCC 8
th
edition, they each have different definitions for T or primary tumor extension. A
schema discriminator is necessary to distinguish between these primary sites so that the appropriate
sub(chapter)/schema is used.
Coding Instructions and Codes
Note: A schema discriminator is used to discriminate between melanoma tumors with primary site code
C694: Ciliary Body/Iris. Code the site in which the tumor arose.
00672: Melanoma Choroid and Ciliary Body (see code 1)
o Subsites include: Ciliary body, crystalline lens, sclera, uveal tract, intraocular, eyeball
00671: Melanoma Iris (see code 2)
o Subsite includes: Iris
Code Description Schema ID#/Description
1 Ciliary Body
Crystalline lens
Sclera
Uveal tract
Intraocular
Eyeball
00672: Melanoma Choroid and Ciliary Body
2 Iris 00671: Iris
Return to Schema ID Table

382 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3821: Chromosome 3 Status
Item Length: 1
NAACCR Item #: 3821
XML Parent-NAACCR ID: Tumor-chromosome3Status
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Chromosome 3 Status refers to the partial or total loss of Chromosome 3, which is a prognostic factor
for uveal melanoma.
Rationale
Chromosome 3 Status is a Registry Data Collection Variable in AJCC. This data item was previously
collected as Uveal Melanoma, CS SSF #5.
Definition
The loss of an entire copy of chromosome 3, which occurs in about half of patients, is the most
important indicator of poor prognosis for the uveal melanomas, particularly melanoma of the choroids
and ciliary body. A variety of sophisticated tests can be used to determine chromosome 3 status
Karyotyping
Fluorescence in situ hybridization
Comparative genomic hybridization
DNA polymorphism analysis (e.g., single nucleotide polymorphism, microsatellite)
Multiplex ligation probe amplification
Monosomy 3 means loss of chromosome 3. Determination of chromosome status may be
affected by prior irradiation
Coding guidelines
Code 0 when there is no loss of chromosome 3, or disomy 3
Code 1 when there is partial loss of chromosome 3
Code 2 when there is complete loss of chromosome 3, or monosomy 3
Code 3 when there is loss of chromosome 3, how much not known
Code 7 when test done, but test results not available
Code 9 when
o No documentation in the medical record
o Chromosome 3 not evaluated (assessed)
o Unknown if Chromosome 3 evaluated (assessed)
o Patients received radiation therapy prior to testing

383 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Source documents: pathology report, specialty/reference lab report, gene expression profile
report, other statement in medical record
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma
Other names: Monosomy 3, loss of chromosome 3, chromosome 3 loss of heterozygosity (LOH),
isodisomy 3 (rare)
Coding Instructions and Codes
Note 1: Physician statement of chromosome 3 status can be used to code this data item when no other
information is available.
Note 2: Monosomy 3, especially if combined with a frequently coexisting gain in chromosome 8q, is
independently associated with metastatic risk. Chromosome 3 and 8 statuses may be determined with
karyotyping or fluorescent in situ hybridization (FISH).
Note 3: See also 3822: Chromosome 8q Status.
Code Description
0 No loss of chromosome 3
1 Partial loss of chromosome 3
2
Complete loss of chromosome 3
3 Loss of chromosome 3, NOS
7 Test ordered, results not in chart
8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Chromosome 3 status not assessed or unknown if assessed
Return to Schema ID Table

384 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3822: Chromosome 8q Status
Item Length: 1
NAACCR Item #: 3822
XML Parent-NAACCR ID: Tumor-chromosome8qStatus
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Chromosome 8q Status refers to gain in Chromosome 8q, which is a prognostic factor for uveal
melanoma.
Rationale
Chromosome 8q Status is a Registry Data Collection Variable in AJCC. This data item was previously
collected as Uveal Melanoma, CS SSF #7.
Definition
The loss of an entire copy of chromosome 8, which occurs in about half of patients, is the most
important indicator of poor prognosis for the uveal melanomas, particularly melanoma of the choroids
and ciliary body. A variety of sophisticated tests can be used to determine chromosome 8 status
Karyotyping;
Fluorescence in situ hybridization;
Comparative genomic hybridization
DNA polymorphism analysis (e.g., single nucleotide polymorphism, microsatellite);
Multiplex ligation probe amplification;
Monosomy 3 means loss of chromosome 3. Determination of chromosome status may be
affected by prior irradiation.
Coding guidelines
Code 0 when there is no gain in chromosome 8q
Code 1 when there is gain in chromosome 8q
Code 7 when test done, but results not available
Code 9 when
o No documentation in the medical record
o Chromosome 8q not evaluated (assessed)
o Unknown if Chromosome 8q evaluated (assessed)
o Patients received radiation therapy prior to testing
Additional Information
Source documents: pathology report, specialty/reference lab report, gene expression profile
report, other statement in medical record

385 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma
Other names: 8q duplication, 8q trisomy, duplication 8q, partial trisomy 8q, trisomy 8q
Coding Instructions and Codes
Note 1: Physician statement of chromosome 8q status can be used to code this data item when no other
information is available.
Note 2: Monosomy 3, especially if combined with a frequently coexisting gain in chromosome 8q, is
independently associated with metastatic risk. Chromosome 3 and 8 statuses may be determined with
karyotyping or fluorescent in situ hybridization.
Note 3: See also 3821: Chromosome 3 Status.
Code Description
0 No gain in chromosome 8q
1
Gain in chromosome 8q
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9
Not
documented in medical record
Chromosome 8q status not assessed or unknown if assessed
Return to Schema ID Table

386 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3834: Extravascular Matrix Patterns
Item Length: 1
NAACCR Item #: 3834
XML Parent-NAACCR ID: Tumor-extravascularMatrixPatterns
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Extravascular Matrix Patterns, the presence of loops and networks in extracellular matrix patterns, is a
prognostic factor for uveal melanoma.
Rationale
Extravascular Matrix Patterns is a Registry Data Collection Variable in AJCC 8. This data item was
previously collected as Uveal Melanoma, CS SSF #11 and CS SSF #12. These two data items were
combined into one data for cases diagnosed 1/1/2018+.
Definition
The presence of extravascular matrix patterns is an indicator for shorter survival. There are two
different types of patterns: loops only, or loops forming networks. The identification of the complex
monocirculatory patterns (i.e., loops, networks, arcs with branching, parallel with cross-linking or a
combination of these patterns) are done using confocal indocyanine green angiography. The patterns
are assessed with light microscopy under a dark green filter after staining with periodic-acid Schiff
without counterstain. This determines the presence or absence of each matrix pattern, which appear
deep purple against a pink background.
Coding guidelines
Code 0 when pathology report states loops, and networks not found
Code 1 when pathology reports states networks and/or loops present
Code 9 when the ER is
o Pathology report available and there is no mention of extravascular matrix patterns
(loops or networks)
o Extravascular matrix patterns not assessed or unknown if assessed
Additional Information
Source documents: pathology report, confocal indocyanine green angiography report, clinician
comment
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma

387 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding instructions and Codes
Note 1: Physician statement of extravascular matrix patterns can be used to code this data item when
no other information is available.
Note 2: The presence of certain types of extravascular matrix patterns is independently associated with
the risk of metastasis. This is documented conclusively for individual loops and for loops forming
networks consisting of at least three back-to-back loops. Absence of both loops and networks is
associated with the longer survival and presence of loops forming networks is associated with the
shortest survival time.
Code Description
0 Extravascular matrix patterns not present/not identified
1 Extravascular matrix patterns present/identified
8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
9 Not documented in medical record
Extravascular Matrix Patterns not assessed or unknown if assessed
Return to Schema ID Table

388 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3887: Measured Basal Diameter
Item Length: 4
NAACCR Item #: 3887
XML Parent-NAACCR ID: Tumor-measuredBasalDiameter
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Measured Basal Diameter, the largest basal diameter of a uveal melanoma, is a prognostic indicator for
this tumor.
Rationale
Measured Basal Diameter is listed as a Registry Data Collection Variable in AJCC. It was previously
collected as Uveal Melanoma, CS SSF #2.
Definition
The basal diameter is the width (horizontal measurement) of the melanoma at its base (in contact with
sclera). This is not the same as the depth of invasion (see NAACCR Data Item #3888-Measured
Thickness). Clinical research has shown that as a uveal tumor becomes larger, the risk of hematogenous
metastases and death increases. In addition, knowing the size of the melanoma is important for
treatment planning.
Per the CAP guidelines for Uveal Melanoma, “in clinical practice, the largest tumor basal diameter may
be estimated in optic disc diameters (dd, average: 1 dd = 1.5 mm). Techniques such as ultrasonography
and fundus photography are used to provide more accurate measurement. When histopathological
measurements are recorded after fixation, tumor diameter and thickness may be underestimated
because of tissue shrinkage.”
Additional Information
Source documents: high-frequency ultrasonography (ultrasound biomicroscopy) report,
pathology report, wide-angle fundus camera measurement, clinician report or other
documentation in medical record
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma
Other names: largest tumor diameter (LTD), tumor basal size; do not code tumor basal area
(measured in square millimeters)

389 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Coding Instructions and Codes
Note 1: Physician statement of measured basal diameter (not the same as tumor size) can be used to
code this data item when no other information is available.
Note 2: Code Measured Basal Diameter of tumor not size. Record actual measurement in millimeters
(mm) to nearest tenth from clinical documentation, or from a pathology report if surgery performed.
Code
Description
0.0 No mass/tumor found
0.1-99.9 0.1 – 99.9 millimeters (mm)
(Exact measurement to nearest tenth of mm)
XX.0 100 millimeters (mm) or larger
XX.1 Described as "less than 3 mm"
XX.2 Described as “at least” 3 mm
XX.3 Described as “at least” 6 mm
XX.4 Described as “at least” 9 mm
XX.5
Described as “at
least” 12 mm
XX.6 Described as “at least” 15 mm
XX.7
Described as “at least” 18 mm
XX.8 Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XX.8 may result in an
edit error.)
XX.9 Not documented in medical record
Cannot be determined by pathologist
Measured Basal Diameter not assessed or unknown if assessed
Return to Schema ID Table

390 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3888: Measured Thickness
Item Length: 4
NAACCR Item #: 3888
XML Parent-NAACCR ID: Tumor-measuredThickness
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Measured Thickness, or height, the thickness of a uveal melanoma, is a prognostic indicator for this
tumor.
Rationale
Measured Thickness is listed as a Registry Data Collection Variable in AJCC. It was previously collected as
Uveal Melanoma, CS SSF #3.
Definition
This data items measures tumor thickness, height or depth (vertical dimension), rather than size (lateral
dimension) of basal diameter (horizontal dimension). (For basal diameter, see NAACCR Data Item #3887-
Measured Basal Diameter).
The depth of invasion or tumor thickness measurement for melanomas of the choroid, ciliary body, and
iris is collected in tenths of millimeters as stated in the pathology report for the resected specimen. (This
is similar to, but not the same as, Breslow depth of invasion, which is measured in hundredths of
millimeters.) Code a measurement specifically labeled as “thickness” “height” or “depth” in the
pathology report. In the absence of this label, a measurement described as taken from the cut surface of
the specimen can be coded. And in the absence of either of these labels, the third dimension in a
statement of tumor size (length x width x depth) can be used by the registrar to code this field.
Per the CAP guidelines for Uveal Melanoma, “in clinical practice, tumor thickness may be estimated in
diopters (average: 2.5 diopters = 1 mm). Techniques such as ultrasonography and fundus photography
are used to provide more accurate measurement. When histopathological measurements are recorded
after fixation, tumor diameter and thickness may be underestimated because of tissue shrinkage.”
Additional Information
Source documents: high-frequency ultrasonography (ultrasound biomicroscopy) report,
pathology report, wide-angle fundus camera measurement, clinician report or other
documentation in medical record
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma
Other names: maximum tumor thickness, depth of invasion; perpendicular tumor diameter

391 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
(PTD); tumor height
Coding Instructions and Codes
Note 1: Physician statement of measured thickness, or height, can be used to code this data item when
no other information is available.
Note 2: Code Measured Thickness, or height, of tumor, not size. Record actual measurement in
millimeters (mm) from clinical documentation, or from a pathology report if surgery performed.
Code
Description
0.0 No mass/tumor found
0.1-99.9 0.1 – 99.9 millimeters (mm)
(Exact measurement to nearest tenth of mm)
XX.0 100 millimeters (mm) or larger
XX.1 Described as "less than 3 mm"
XX.2 Described as “at least” 3 mm
XX.3 Described as “at least” 6 mm
XX.4 Described as “at least” 9 mm
XX.5 Described as “at least” 12 mm
XX.6
Described as “greater than” 15 mm
XX.8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XX.8 may result in an
edit error.)
XX.9 Not documented in medical record
Cannot be determined
Measured Thickness not assessed or unknown if assessed
Return to Schema ID Table

392 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3891: Microvascular Density
Item Length: 2
NAACCR Item #: 3891
XML Parent-NAACCR ID: Tumor-microvascularDensity
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Microvascular Density, a quantitative measure of tumor vascularity, is a prognostic factor for uveal
melanoma.
Rationale
Microvascular Density, is a Registry Data Collection Variable in AJCC. This data item was previously
collected as Uveal Melanoma, CS SSF #13.
Definition
A high density of microvessels, identified immunohistochemically using antibodies for vascular
endothelial cells (such as Factor VIII-related antigen, CD34 epitope, etc.), has prognostic significance in a
melanoma of the uvea. Higher counts have more unfavorable outcome. To obtain microvascular density,
the pathologist, using a microscope with an eyepiece graticule (grid) of approximately 0.3 square mm
and X200 magnification, counts microvessels from the most highly vascularized areas (“hot spots”) of
the tumor, identified by scanning the entire immunostained tumor at lower magnification. Any
immunolabeled element, clearly separate from an adjacent one and either totally inside the graticule or
touching its top or left border, is counted as a microvessel. In several studies, the range of microvascular
density was from 5 to 121 vessels, although this will vary depending on the type of immunostaining and
area of graticule used.
Code the microvascular density (number of microvessels) in whole numbers as stated in the pathology
report in the code range 001 (1 vessel per 0.3 square millimeters) to 500 (500 vessels per 0.3 square
millimeters).
Additional Information
Source documents: pathology report
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma
Coding Instructions and Codes
Note 1: Physician statement of microvascular density (MVD) can be used to code this data item when no
other information is available.

393 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 2: MVD is independently associated with metastatic risk. The number of immunopositive elements
is labeled with a marker for vascular endothelial cells (e.g., CD34 epitope, CD31 epitope, factor VIII-
related antigen) and counted from area of densest vascularization (typical field area, 0.3 mm2 squared).
Higher counts are associated with shorter survival.
Note 3: Record the results as expressed on the laboratory test. Record the information based on
quartiles for laboratory standards if this is the only expression of results.
Code Description
00 No vessels involved
01-99 01-99 vessels per 0.3 square millimeter (mm2)
X1 Greater than or equal to 100 vessels per 0.3 square millimeter (mm2)
X2 Lowest quartile for laboratory
X3 Second quartile for laboratory
X4 Third quartile for laboratory
X5 Highest quartile for laboratory
X7 Test ordered, results not in chart
X8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code 8 may result in an edit
error.)
X9
Not documented in medical record
Microvascular Density (MVD) not assessed or unknown if assessed
Return to Schema ID Table

394 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00671: Melanoma Iris (2018+)
3892: Mitotic Count Uveal Melanoma
Item Length: 4
NAACCR Item #: 3892
XML Parent-NAACCR ID: Tumor-mitoticCountUvealMelanoma
NAACCR Alternate Name: Mitotic None
Active years: 2018+
Schema(s):
00671: Melanoma Iris (2018+)
00672: Melanoma Choroid and Ciliary Body (2018+)
Description
Mitotic Count Uveal Melanoma, the number of mitoses per 40 high-power fields (HPF) based on
pathological evaluation, is a prognostic factor for uveal melanoma.
Rationale
Mitotic Count Uveal Melanoma is listed as a Registry Data Collection Variable in AJCC. It was previously
collected as Uveal Melanoma, CS SSF #9.
Definition
Mitotic count is collected for several different types of cancers. For melanomas of the choroids, ciliary
body and iris, the standard measurement is the total number of mitoses per 40 high power fields (HPF at
40 times magnification) per 0.152 square millimeters.
Additional Instructions
Source documents: pathology report
For further information, refer to the Uveal Melanoma cancer protocol published by the College
of American Pathologists for the AJCC Staging System Uveal Melanoma
Coding Instructions and Codes
Note 1: Physician statement of mitotic count for a uveal melanoma can be used to code this data item
when no other information is available.
Note 2: The mitotic count, the number of mitoses per 40 high-power fields (HPF), reflects the potential
aggressiveness or prognosis of uveal melanomas. This data item presumes the denominator of 40 HPF,
so just the numerator (the mitotic count) is coded here.
For other schemas in which mitotic count is collected, the denominator may vary.
Note 3: An HPF usually has a magnification objective of 40 (a 40x field). As described in the AJCC
chapter on uveal melanomas, the typical field area is 0.152 square millimeters (mm2).
Note 4: Record mitotic count to the nearest tenth as documented in the pathology report.
For example, a mitotic count of 6/40 HPF would be coded 6.0.

395 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
0.0 0 mitoses per 40 high-power fields (HPF)
Mitoses absent, no mitoses present, no mitotic activity
0.1-99.9 0.1-99.9 mitosis per 40 HPF
XX.1 100 or more mitoses per 40 HPF
XX.2 Stated as low mitotic count or rate with no specific number
XX.3 Stated as high mitotic count or rate with no specific number
XX.4 Mitotic count described with denominator other than 40 HPF
XX.7
Test
ordered
, results not in chart
XX.8
Not applicable: Information not collected for this case
(If this information is required by your standard setter, use of code XX.8 may result in an
edit error.)
XX.9 Not documented in medical record
Mitotic Count Uveal Melanoma not assessed or unknown if assessed
Return to Schema ID Table

396 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00672: Melanoma Choiroid and Ciliary Body (2018+)
See 00671: Melanoma Iris
3926: Schema Discriminator 1: Melanoma Ciliary Body/Melanoma Iris
3821: Chromosome 3 Status
3822: Chromosome 8q Status
3834: Extravascular Matrix Patterns
3887: Measured Basal Diameter
3888: Measured Thickness
3891: Microvascular Density
3892: Mitotic Count Uveal Melanoma

397 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00680: Retinoblastoma (2018+)
3856: Heritable Trait
Item Length: 1
NAACCR Item #: 3856
XML Parent-NAACCR ID: Tumor-heritableTrait
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00680: Retinoblastoma (2018+)
Description
Heritable trait pertains to evidence that a tumor is associated with a heritable mutation. In
retinoblastoma, the heritable trait is a germline mutation in the RB1 gene, which is associated with
bilateral disease, family history of retinoblastoma, presence of concomitant CNS midline embryonic
tumor (commonly in pineal region), or retinoblastoma with an intracranial primitive neuroectodermal
tumor (i.e., trilateral retinoblastoma). Children with any of these features may be assigned the H1 status
without molecular testing. High quality molecular testing for RB1 mutation is required to determine the
presence or absence of RB1 mutation for children without clinical features of a heritable mutation.
Rationale
Heritable trait is required for prognostic stage grouping in AJCC 8
th
edition, Chapter 68 Retinoblastoma.
It is a new data item for cases diagnosed 1/1/2018+.
Definition
Heritable disease (trait) is defined by the presence of a germline mutation of the RB1 gene. This
germline mutation may have been inherited from an affected progenitor (25% of cases) or may have
occurred in a germ cell before conception or in utero during early embryogenesis in patients with
sporadic disease (75% of cases). The presence of positive family history or bilateral or multifocal disease
is suggestive of heritable disease.
Heritable retinoblastoma may manifest as unilateral or bilateral disease. The penetrance of the RB1
mutation (laterality, age at diagnosis, and number of tumors) is probably dependent on concurrent
genetic modifiers such as MDM2 and MDM4 polymorphisms. All children with bilateral disease and
approximately 15% of patients with unilateral disease are presumed to have the heritable form, even
though only 25% have an affected parent.
In heritable retinoblastoma, tumors tend to be diagnosed at a younger age than in the nonheritable
form of the disease. Unilateral retinoblastoma in children younger than 1 year raises concern for
heritable disease, whereas older children with a unilateral tumor are more likely to have the
nonheritable form of the disease.
Children with a germline RB1 mutation may continue to develop new tumors for a few years after
diagnosis and treatment; for this reason, they need to be examined frequently. It is common practice for

398 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
examinations to occur every 2 to 4 months for at least 28 months. The interval between exams is based
on the stability of the disease and age of the child (i.e., less frequent visits as the child ages).
Patients with heritable retinoblastoma are also at a greater risk for subsequent neoplasms.
Heritable trait is required for prognostic stage grouping in the AJCC Staging System Retinoblastoma. It is
a new data item for cases diagnosed 1/1/2018+.
Additional Information
Source documents: lab reports (blood), pathology report
Coding Instructions and Codes
Note 1: Physician statement of retinoblastoma heritable trait can be used to code this data item.
Note 2: Code Heritable trait (H) based on the criteria listed in Chapter 68 Retinoblastoma “Definition of
Heritable Trait (H).”
Note 3: Code 0 (H0) if clinical features do not exist or laboratory germline RB1 test is negative or there is
no clinical evidence of mutation. Results may be from blood or tissue testing.
Note 4: Code 0 (H0) if residual (false negative) risk for a mutation is less than 1% or at population risk
(0.007%) in a laboratory with demonstrated sensitivity greater than 97%.
Note 5: Code 1 (H1) may be assigned based on positive molecular testing for germline RB1 gene.
Note 6: Code 1 (H1) may be assigned based on clinical evidence of any of the following features even
without molecular testing (in particular for children). When discrete clinical evidence of heritable trait is
not present, high-quality molecular evidence is mandatory before designating a child as H1 positive.
Bilateral disease
Family history of retinoblastoma
Presence of concomitant CNS midline embryonic tumor (commonly in pineal region)
Retinoblastoma with an intracranial primitive neuroectodermal tumor (i.e., trilateral
retinoblastoma)
Note 7: Variants of unknown significance should be categorized as 9 (HX).
Code Description
0 H0: Normal RB1 alleles
No clinical evidence of mutation
1 H1: RB1 gene mutation OR
Clinical evidence of mutation
7 Test ordered, results not in chart
9 HX: Not documented in medical record
Test not done, or unknown if done
Insufficient evidence of a constitutional RB1 gene mutation
Return to Schema ID Table

400 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00690: Lacrimal Gland (2018+)
3926: Schema Discriminator 1: Lacrimal Gland/Sac
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00690: Lacrimal Gland (2018+)
00698: Lacrimal Duct (2018+)
Definition
The lacrimal (also spelled lachrymal) gland is the only epithelial structure normally present within the
orbit. Its composition is the same as epithelial salivary glands and AJCC TNM staging parallels that of the
major salivary gland classification
Lacrimal gland and lacrimal sac have the same ICD-O topography code (C695). However, for purposes of
the AJCC Staging System stage grouping, lacrimal gland is AJCC staged while lacrimal sac is not (Summary
Stage only). A schema discriminator is necessary to distinguish between these primary sites so that the
appropriate system/schema is used.
Coding Instructions and Codes
Note 1: A schema discriminator is used to discriminate between lacrimal gland and lacrimal sac tumors
with primary site code C695: Lacrimal Gland. Code the site in which the tumor arose.
Note 2: If the histology is transitional cell carcinoma (8120/3, 8130/3), assign code 2.
00690: Lacrimal Gland (see code 1)
o Subsites include: lacrimal gland
00698: Lacrimal Sac (see code 2)
o Subsites include: lacrimal sac, lacrimal duct (NOS), nasal lacrimal duct
Code Description Schema ID#/Description
1 Lacrimal gland 00690: Lacrimal Gland
2
Lacrimal sac
Lacrimal duct, NOS
Nasal lacrimal duct/sac
Nasolacrimal duct
00698: Lacrimal Sac
9 Lacrimal, NOS 00698: Lacrimal Sac
Return to Schema ID Table

401 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00690: Lacrimal Gland (2018+)
3803: Adenoid Cystic Basaloid Pattern
Item Length: 5
NAACCR Item #: 3803
XML Parent-NAACCR ID: Tumor-adenoidCysticBasaloidPattern
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00690: Lacrimal Gland (2018+)
Description
Adenoid Cystic Basaloid Pattern, the presence of a basaloid pattern on pathological examination, is a
prognostic factor for adenoid cystic carcinoma of the lacrimal gland.
Rationale
Adenoid Cystic Basaloid Pattern is a Registry Data Collection Variable in AJCC 8. This data item was
previously collected as Lacrimal Gland, CS SSF #6.
Definition
Adenoid cystic carcinoma (ICD-O-3 morphology code 8200/3) is the most common malignant epithelial
tumor of the lacrimal gland. Adenoid cystic carcinoma is a tumor composed of modified myoepithelial
and ductal differentiated cells. A genetic alteration (i.e., fusion oncogene MYB-NFIB) is found in the
majority of adenoid cystic carcinomas There are three histologic patterns within the adenoid cystic
carcinoma group: cribriform, solid and tubular.
Coding guidelines
Code 0.0 when the pathology report states that basaloid or solid pattern is not present
Code 0.1-100.0 when the pathology report states the percent of basaloid or solid pattern that is
present
Code XXX.5 when basaloid or solid pattern present but percentage not known;
Code XXX.9 when
o Histopathologic pattern not documented in the medical record
o Histopathologic pattern not evaluated (assessed)
o Unknown if histopathologic pattern evaluated (assessed)
o When histologic type other than 8200 and there is no mention of basaloid pattern (see
Note 2 under coding instructions)
Additional Information
o Source documents: pathology report
o Other names: ACC, basaloid type adenoid cystic carcinoma
Coding Instructions and Codes

402 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 1: Physician statement of basaloid pattern can be used to code this data item when no other
information is available.
Note 2: This is most commonly found in Adenoid Cystic Carcinoma (8200/3) but can be present in other
histologies.
Code
Description
0.0-100.0 0.0 to 100.0 percent basaloid pattern
XXX.5 Basaloid pattern present, percentage not stated
XXX.8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code XXX.8 will result in an edit
error.)
XXX.9 Not documented in medical record
Adenoid Cystic Basaloid Pattern not assessed or unknown if assessed
Return to Schema ID Table
404 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
CENTRAL NERVOUS SYSTEM

405 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00721: Brain (2018-2022)
3816: Brain Molecular Markers
Item Length: 2
NAACCR Item #: 3816
XML Parent-NAACCR ID: Tumor-brainMolecularMarkers
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00721: Brain (2018-2022)
09721: Brain (2023+)
00722: CNS Other (2018-2022)
09722: CNS Other (2023+)
Description
Multiple brain molecular markers have become standard pathology components necessary for
diagnosis. This data item captures clinically important brain cancer subtypes identified by molecular
markers that are not distinguishable by ICD-O-3 codes.
Rationale
Collection of these clinically important brain cancer subtypes has been recommended by CBTRUS.
Coding Instructions and Codes
Note 1: This data item applies only to ICD-O-3 histology codes: 9400/3, 9401/3, 9440/3, 9450/3,
9451/3, 9471/3 and 9478/3. If a microscopically confirmed histology is not included in this list, assign,
code 85.
If your case is not microscopically confirmed, code 99
Note 2: Physician statement of histologic subtype can be used to code this data item.
Note 3: Only one code is applicable for each tumor.
IDH mutation status distinguishes between clinically important subtypes within ICD-O-3 9400/3,
Diffuse astrocytoma and 9401/3, Anaplastic astrocytoma.
IDH mutant and 1p/19q co-deletion distinguishes between clinically important subtypes within
ICD-O-3 code 9450/3, Oligodendroglioma and 9451/3, Anaplastic Oligodendroglioma.
IDH-wildtype distinguishes clinically important subtypes within ICD-O-3 9400/3, Diffuse
astrocytoma, 9401/3, Anaplastic astrocytoma and 9440/3, Glioblastoma, Epithelioid
glioblastoma and Glioblastoma, NOS (note that the new ICD-O-3 code 9445/3 applies to
Glioblastoma, IDH-mutant; information regarding this subtype is not collected using this data
item).
SHH-activation and TP53-wildtype distinguishes between clinically important subtypes within
ICD-O-3 histology code 9471/3, Medulloblastoma.
C19MC alteration status distinguishes a clinically important highly aggressive subtype within
ICD-O-3 9478/3, Embryonal tumor with multilayered rosettes.
Examples:

406 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
1. Biopsy of brain tumor, microscopic confirmation diagnosis: Diffuse Astrocytoma (9400/3). Additional
testing done, and IDH-mutant is identified. Code 01. Biopsy of brain tumor, microscopic
confirmation diagnosis: Anaplastic astrocytoma (9401/3). No further testing or results unknown.
Code 99.
2. MRI of brain tumor, clinical diagnosis: glioblastoma. No further workup. Code 99.
3. Biopsy of brain tumor, microscopic confirmation diagnosis: Mixed glioma (9382/3). Code 85.
Code
Description
01 Diffuse astrocytoma, IDH-mutant (9400/3)
02 Diffuse astrocytoma, IDH-wildtype (9400/3)
03 Anaplastic astrocytoma, IDH-mutant (9401/3)
04 Anaplastic astrocytoma, IDH-wildtype (9401/3)
05 Glioblastoma, IDH-wildtype (9440/3)
06 Oligodendroglioma, IDH-mutant and 1 p/19q co-deleted (9450/3)
07 Anaplastic oligodendroglioma, IDH-mutant and 1p/19q co-deleted (9451/3)
08 Medulloblastoma, SHH-activated and TP53-wildtype (9471/3)
09 Embryonal tumor with multilayered rosettes, C19MC-altered (9478/3)
85 Not applicable: Histology not 9400/3, 9401/3, 9440/3, 9450/3, 9451/3,
9471/3, 9478/3
86 Benign or borderline tumor
87 Test ordered, results not in chart
88 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 88 will result in an
edit error.)
99 Not documented in medical record
No microscopic confirmation
Brain molecular markers not assessed or unknown if assessed
Return to Schema ID Table

407 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00721: Brain (2018-2022)
3801, 3802: Loss of Heterozygosity: Chromosome 1p and Chromosome 19q (CNS)
Definition
These two genetic tests are frequently done at the same time and reported together. Loss of
heterozygosity (LOH) in a chromosome means that genetic material normally found in a specific area of
a chromosome is missing. In other words, this is damage to the chromosome that results in failure of
tumor suppression, which in turn may cause the development or progression of a malignancy. For 1p
LOH, the specific chromosomal defect is on the short arm (p) of chromosome 1. For 19q LOH, the
specific chromosomal defect is on the long arm (q) of chromosome 19.
Normal cells have two complete copies of each chromosome, a state called heterozygosity. The loss of
this section of the chromosome is associated with improved outcome. It can be used to aid diagnosis
and to make treatment decisions because sensitivity to chemotherapy agents, such as lomustine,
procarbazine, and vincristine, is increased with either 1p or 19q LOH. Special molecular diagnostic
(polymerase chain reaction or gene amplification) tests look for missing genetic material. LOH for
chromosome 1p and 19q is tested primarily for oligodendroglioma, anaplastic oligodendroglioma,
oligoastrocytoma, and anaplastic oligoastrocytoma. It is infrequently tested for other gliomas, such as
glioblastoma multiforme.
Coding guidelines
Code 0 when the 1p/19q is not identified/not present
Code 1 when the 1p/19q is present
Code 7 when the 1p/19q test was ordered but the results are not in the medical record
Code 9 when
o No documentation in the medical record
o 1p/19q test not done (not assessed)
o Unknown if 1p/19q test was performed (unknown if assessed)
Additional Information
Other names allelic loss, gene deletion, 1p/19q fragment analysis
Return to Schema ID Table

408 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00721: Brain (2018-2022)
3801: Chromosome 1p: Loss of Heterozygosity (LOH)
Item Length: 1
NAACCR Item #: 3801
XML Parent-NAACCR ID: Tumor-chromosome1pLossHeterozygosity
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00721: Brain (2018-2022)
09721: Brain (2023+)
00722: CNS Other (2018-2022)
09722: CNS Other (2023+)
Description
Chromosome 1p: Loss of Heterozygosity (LOH) refers to the loss of genetic material normally found on
the short arm of one of the patient's two copies of chromosome 1. Codeletion of Chromosome 1p and
19q is a diagnostic, prognostic and predictive marker for gliomas and is strongly associated with the
oligodendroglioma phenotype.
Rationale
Chromosome 1p: Loss of Heterozygosity (LOH) is a Registry Data Collection Variable in AJCC. It was
previously collected as Brain, CS SSF #5.
See 3801, 3802: Loss of Heterozygosity: Chromosome 1p and Chromosome 19q (CNS) for additional
information.
Coding Instructions and Codes
Note 1: Physician statement of Chromosome 1p deletion/LOH can be used to code this data item.
Note 2: This is a special molecular diagnostic test performed on tumor tissue to identify loss of genetic
material normally found on the short arm of one of the patient's two copies of chromosome 1. A normal
cell will contain two complete copies of each chromosome, one from each parent, and this normal state
is termed heterozygous. Loss of heterozygosity (LOH) is an abnormal state reflecting loss of the whole
arm of chromosome 1p following a chromosomal translocation event.
Note 3: Other terms for LOH include whole arm loss, gene deletion and allelic loss.
Note 4: Below is a list of histologies/terms for which the Chromosome 1p test is commonly done. If the
test was done, record the results, regardless of the histology.
9382/3: Oligoastrocytoma (anaplastic, or NOS)
9400/3: Diffuse astrocytoma (IDH mutant, IDH wild type, or NOS)
9401/3: Anaplastic astrocytoma (IDH mutant, IDH wild type, or NOS)
9411/3: Gemistocytic astrocytoma, IDH mutant
9424/3: Anaplastic pleomorphic xanthoastrocytoma
9430/3: Astroblastoma
9440/3: Glioblastoma (epithelioid, IDH wild type, or NOS)

409 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
9441/3: Giant cell glioblastoma
9442/3: Gliosarcoma
9445/3: Glioblastoma, IDH mutant
9450/3: Oligodendroglioma (IDH mutant and 1p/19q codeleted, or NOS)
9451/3: Anaplastic oligodendroglioma (IDH mutant and 1p/19q codeleted, or NOS)
9505/3: Anaplastic ganglioglioma
9530/3: Anaplastic (malignant) meningioma
Note 5: If the histology is not listed among those for which the Chromosome 1p test is commonly done,
and the test result is not readily available, assume it was not done and code 9 for unknown.
Note 6: For brain tumors, tests for LOH of chromosomes 1p and 19q may be performed at the same
time and reported on a single report. See also 3802: Chromosome 19q: Loss of Heterozygosity (LOH).
Code Description
0 Chromosome 1p deletion/LOH not identified/not present
1 Chromosome 1p deletion/LOH identified/present
6 Benign or borderline tumor
7 Test ordered, results not in chart
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Cannot be determined by the pathologist
Chromosome 1p deletion/LOH not assessed or unknown if assessed
Return to Schema ID Table

410 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00721: Brain (2018-2022)
3802: Chromosome 19q: Loss of Heterozygosity (LOH)
Item Length: 1
NAACCR Item #: 3802
XML Parent-NAACCR ID: Tumor-chromosome19qLossHeterozygosity
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00721: Brain
00722: CNS Other
Description
Chromosome 19q: Loss of Heterozygosity (LOH) refers to the loss of genetic material normally found on
the long arm of one of the patient's two copies of chromosome 19. Codeletion of Chromosome 1p and
19q is a diagnostic, prognostic and predictive marker for gliomas and is strongly associated with the
oligodendroglioma phenotype.
Rationale
Chromosome 19q: Loss of Heterozygosity (LOH) is a Registry Data Collection Variable in AJCC. It was
previously collected as Brain, CS SSF #6.
See 3801, 3802: Loss of Heterozygosity: Chromosome 1p and Chromosome 19q (CNS) for additional
information.
Coding Instructions and Codes
Note 1: Physician statement of Chromosome 19q deletion/LOH can be used to code this data item.
Note 2: This is a special molecular diagnostic test performed on tumor tissue to identify loss of genetic
material normally found on the long arm of one of the patient's two copies of chromosome 19. A normal
cell will contain two complete copies of each chromosome, one from each parent, and this normal state
is termed heterozygous. Loss of heterozygosity (LOH) is an abnormal state reflecting loss of the whole
arm of chromosome 19q following a chromosomal translocation event.
Note 3: Other terms for LOH include whole arm loss, deletion and allelic loss.
Note 4: Below is a list of histologies/terms for which the Chromosome 19q test is commonly done. If the
test was done, record the results, regardless of the histology.
9382/3: Oligoastrocytoma (anaplastic, or NOS)
9400/3: Diffuse astrocytoma (IDH mutant, IDH wild type, or NOS)
9401/3: Anaplastic astrocytoma (IDH mutant, IDH wild type, or NOS)
9411/3: Gemistocytic astrocytoma, IDH mutant
9424/3: Anaplastic pleomorphic xanthoastrocytoma
9430/3: Astroblastoma
9440/3: Glioblastoma (epithelioid, IDH wild type, or NOS)
9441/3: Giant cell glioblastoma
9442/3: Gliosarcoma

411 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
9445/3: Glioblastoma, IDH mutant
9450/3: Oligodendroglioma (IDH mutant and 1p/19q codeleted, or NOS)
9451/3: Anaplastic oligodendroglioma (IDH mutant and 1p/19q codeleted, or NOS)
9505/3: Anaplastic ganglioglioma
9530/3: Anaplastic (malignant) meningioma
Note 5: If the histology is not listed among those for which the Chromosome 1p test is commonly done,
and the test result is not readily available, assume it was not done and code 9 for unknown.
Note 6: For brain tumors, tests for LOH of chromosomes 1p and 19q may be performed at the same
time and reported on a single report. See also 3801: Chromosome 1p: Loss of Heterozygosity (LOH).
Code Description
0 Chromosome 19q deletion/LOH not identified/not present
1 Chromosome 19q deletion/LOH present
6 Benign or borderline tumor
7 Test ordered, results not in chart
8
Not applicable:
Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Cannot be determined by the pathologist
Chromosome 19q: LOH not assessed or unknown if assessed
Return to Schema ID Table

412 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00721: Brain (2018-2022)
3889: Methylation of O6-Methylguanine-Methyltransferase
Item Length: 1
NAACCR Item #: 3889
XML Parent-NAACCR ID: Tumor-methylationOfO6MGMT
NAACCR Alternate Name: Methylation of O6-Methylguanine-Methyltransferase (MGMT)
Active years: 2018+
Schema(s):
00721: Brain (2018)
09721: Brain (2023+)
00722: CNS Other
Description
O6-Methylguanine-Methyltransferase (MGMT) is an enzyme in cells that repairs DNA. Methylation of
the MGMT gene reduces production of the MGMT enzyme and the ability of tumor cells to repair
damage caused by chemotherapy. Methylation of MGMT is a prognostic and predictive factor for high
grade gliomas.
Rationale
Methylation of O6-Methylguanine-Methyltransferase (MGMT) is a Registry Data Collection Variable in
AJCC. It was previously collected as Brain, CS SSF #4.
Definition
O6-Methylguanine-Methyltransferase (MGMT) is an enzyme in cells that repairs DNA. DNA repair is
undesirable in tumors, because it may enable them to overcome the DNA damage done by
chemotherapy. With methylation, less MGMT enzyme is produced, which may lead to prolonged
survival compared to unmethylated MGMT
A patient with increased MGMT methylation is more likely to respond to alkylating agents such as
temozolomide (Temodar) and the nitrosoureas, some of the few drugs effective for brain tumors.
MGMT methylation is a special (not routine) molecular test done on tumor tissue. It is used primarily for
anaplastic oligodendroglioma, anaplastic astrocytoma and glioblastoma multiforme, but can also be
done for low grade malignant central nervous system tumors.
Coding guidelines
Code 0 when the MGMT is not identified/not present
Code 1 when the MGMT is low
Code 2 when the MGMT is high
Code 3 when the MGMT is mentioned, but not stated as low or high
Code 6 for a benign (/0) or borderline (/1) tumor
Code 7 when the MGMT test was ordered but the results are not in the medical record.
Code 9 when
o No information in the medical record about MGMT
o MGMT test not done (not assessed)
o Unknown if MGMT test was performed (unknown if assessed)

413 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Source documents: pathology report, specialty or reference laboratory report
Other names: MGMT promoter methylation, methylation status
Coding Instructions and Codes
Note 1: Physician statement of the methylation status of the MGMT, also termed MGMT promoter,
gene can be used to code this data item.
Note 2: O6-Methylguanine-Methyltransferase (MGMT) is an enzyme in cells that repairs DNA. DNA
repair is undesirable in tumors, because it may enable them to overcome the DNA damage done by
chemotherapy. With methylation, less MGMT enzyme is produced, which may lead to prolonged
survival compared to unmethylated MGMT.
Note 3: Below is a list of histologies/terms for which the MGMT test is commonly done. If the test was
done, record the results, regardless of the histology.
9382/3: Anaplastic oligoastrocytoma, NOS
9382/3: Oligoastrocytoma, NOS
9400/3: Diffuse astrocytoma (IDH mutant, IDH wild type, NOS)
9401/3: Anaplastic astrocytoma (IDH mutant, IDH wild type, NOS)
9411/3: Gemistocytic astrocytoma, IDH mutant
9424/3: Anaplastic pleomorphic xanthoastrocytoma
9440/3: Glioblastoma (epithelioid, IDH wild type, NOS)
9441/3: Giant cell glioblastoma
9442/3: Gliosarcoma
9445/3: Glioblastoma, IDH mutant
9450/3: Oligodendroglioma (IDH mutant and 1p/19q codeleted, NOS)
9451/3: Anaplastic oligodendroglioma (IDH mutant and 1p/19 codeleted, NOS)
9505/3: Anaplastic ganglioglioma
9530/3: Anaplastic (malignant)meningioma
Note 4: If the histology is not listed among those for which the MGMT test is commonly done, and the
test result is not readily available, assume it was not done and code 9 for unknown.
Code
Description
0 MGMT methylation absent/not present, unmethylated MGMT
1 MGMT methylation present, low level
Hypomethylated
Partial methylated
2
MGMT methylation present, high level
Hypermethylated
3 MGMT Methylation present, level unspecified
6 Benign or borderline tumor
7 Test ordered, results not in chart
8
Not applicable:
Information not
collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
Cannot be determined by the pathologist
MGMT not assessed or unknown if assessed
Return to Schema ID Table

414 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
09721: Brain (2023+)
See 00721: Brain (2018-2022)
3816: Brain Molecular Markers
3801: Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity (LOH)
3889: Methylation of O6-Methylguanine-Methyltransferase
Return to Schema ID Table

415 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00722: CNS Other (2018-2022)
See 00721: Brain (2018-2022)
3816: Brain Molecular Markers
3801: Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity (LOH)
3889: Methylation of O6-Methylguanine-Methyltransferase
Return to Schema ID Table

416 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
09722: CNS Other (2023+)
See 00721: Brain (2018-2022)
3816: Brain Molecular Markers
3801: Chromosome 1p: Loss of Heterozygosity (LOH)
3802: Chromosome 19q: Loss of Heterozygosity (LOH)
3889: Methylation of O6-Methylguanine-Methyltransferase
Return to Schema ID Table
418 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
ENDOCRINE SYSTEM

419 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00730: Thyroid (2018+)
3926: Schema Discriminator 1: Thyroid Gland/Thyroglossal Duct
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00730: Thyroid (2018+)
00740: Thyroid Medullary (2018+)
Definition
Thyroid, NOS and thyroglossal duct have the same ICD-O topography code (C739). However, for
purposes of the AJCC Staging Systems Thyroid and Thyroid Medullary stage groupings, Thyroid, NOS is
applicable for AJCC staging while thyroglossal duct is not (Summary Stage only). A schema discriminator
is necessary to distinguish between these primary sites so that the appropriate system/schema is used.
Coding Instructions and Codes
Note: A schema discriminator is used to discriminate between thyroid gland and thyroglossal duct
tumors with primary site code C739: Thyroid Gland. Code the site in which the tumor arose.
Thyroid gland (see code 1)
o Subsites include: Thyroid, NOS
Thyroglossal duct (see code 2)
Code Description
1
Thyroid gland
Thyroid, NOS
2 Thyroglossal duct cyst
Return to Schema ID Table
421 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
HEMATOLOGIC MALIGNANCIES

422 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00790: Lymphoma (excluding CLL/SLL) (2018+)
3926: Schema Discriminator 1: Histology Discriminator for 9591/3
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00790: Lymphoma (2018+)
00830: HemeRetic (2018+)
Coding Notes and Instructions
Note: A schema discriminator is used to discriminate for histology 9591/3: Non-Hodgkin lymphoma to
determine which AJCC Stage Group table to use.
9591/3: Splenic B-cell lymphoma/leukemia, unclassifiable (see code 1)
Abstracted and staged as a leukemia
9591/3: Hairy cell leukemia variant (see code 2)
Abstracted and staged as a leukemia
9591/3: Splenic diffuse red pulp small B-cell lymphoma (see code 3)
Abstracted and staged as a lymphoma
9591/3: Non-Hodgkin lymphoma, NOS (see code 9)
Abstracted and staged as a lymphoma
Code Description Schema ID#/Description
1 Splenic B-cell lymphoma/leukemia, unclassifiable 00830: HemeRetic
2
Hairy cell leukemia variant
Prolymphocytic variant of hairy cell leukemia
00830: HemeRetic
3
Splenic diffuse red
pulp small B
-
cell lymphoma
Splenic marginal zone lymphoma, diffuse variant
Splenic red pulp lymphoma with numerous basophilic
villous lymphocytes
Splenic lymphoma with villous lymphocytes
00790: Lymphoma
(excluding CLL/SLL)
9 Non-Hodgkin lymphoma, NOS
Any other terminology describing NHL
00790: Lymphoma
(excluding CLL/SLL)
<Blank> Histology is NOT 9591, Discriminator is not necessary
Return to Schema ID Table

423 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00790: Lymphoma (excluding CLL/SLL) (2018+)
3812: B Symptoms
Item Length: 1
NAACCR Item #: 3812
XML Parent-NAACCR ID: Tumor-bSymptoms
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00790: Lymphoma (excluding CLL/SLL) (2018+)
00795: Lymphoma-CLL/SLL (2018+)
Description
B symptoms refer to systemic symptoms of fever, night sweats, and weight loss which can be associated
with both Hodgkin lymphoma and some non-Hodgkin lymphomas. The presence of B symptoms is a
prognostic factor for some lymphomas.
Rationale
B symptoms is a Registry Data Collection Variable in AJCC. This data item was previously collected for
Lymphomas, SSF #2.
The stages of Hodgkin Lymphoma are classified as either A or B according to the absence or presence of
defined constitutional symptoms. The stage group suffix for a patient without these systemic symptoms
is “A,” meaning absence of symptoms or asymptomatic; for example, Stage IIA. The stage group suffix
for a patient with any of the symptoms listed below is “B,” such as Stage IIIB. The symptoms are
carefully defined
Fevers: Unexplained fever with temperature above 38 degrees centigrade or 101.5 degrees
Fahrenheit.
Night sweats: Drenching sweats (e.g. those that require change of bedclothes)
Weight loss: Unexplained weight loss of more than 10% of the usual body weight in the 6
months prior to diagnosis.
Other symptoms, such as chills, pruritic, alcohol-induced pain and fatigue, are not included in the A
or B designation but are recorded in the medical record, as the reappearance of these symptoms
may be a harbinger of recurrence. The designation A or B is not included in the revised staging of
NHL in AJCC8, although clinicians are encouraged to record the presence of these symptoms in the
medical record. The presence or absence of B symptoms may be collected in registries for both HL
and NHL.
Coding guidelines
Code 0 when there is no evidence of B symptoms present, per physician or physical exam
Code 1 when the physician states the patient has B symptoms
Code 9 when
o Not documented in the medical record
o B symptoms not evaluated (assessed)
o Unknown if B symptoms evaluated (assessed)

424 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Additional Information
Source documents: patient history, progress notes, consultant notes, other statements in
medical record
Other names: B symptoms; Fever: Palestine fever, hyperpyrexia, febrile response; sleep
hyperhidrosis, nocturnal hyperhydrosis
Note: This was previously required for staging under the Ann Arbor Staging Classification for
Lymphomas. The new Lugano Staging System does not require this for staging.
o Per AJCC 8
th
edition: “The designation A or B is not included in the revised staging of
NHL, although clinicians are encouraged to record the presence of these symptoms
in the medical record.”
o If your physicians no longer record the B symptoms because of this change, code 9
Change from Collaborative Stage v2 (CSv2): In CSv2, if there was no mention of B
symptoms, the registrar could assume that they were not present and code appropriately.
For the SSDI, this assumption cannot be made. There must be a statement that B symptoms
are not present to assign code 0.
Coding Instructions and Codes
Note 1: Physician statement of B symptoms can be used to code this data item when no other
information is available.
Note 2: Each stage should be classified as either A or B according to the absence or presence of defined
constitutional symptoms, such as
Fevers: Unexplained fever with temperature above 38 degrees C
Night sweats: Drenching sweats that require change of bedclothes
Weight loss: Unexplained weight loss of more than 10% of the usual body weight in the six
months prior to diagnosis
Note 3: Pruritus alone does not qualify for B classification, nor does alcohol intolerance, fatigue, or a
short, febrile illness associated with suspected infections.
Note 4: Code 9 if there is no mention of B symptoms.
Code Description
0 No B symptoms (asymptomatic)
Classified as “A” by physician when asymptomatic
1 Any B symptom(s)
Night sweats (drenching)
Unexplained fever (above 38 degrees C)
Unexplained weight loss (generally greater than 10% of body weight
in the six months before admission)
B symptoms, NOS
Classified as “B” by physician when symptomatic
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
B symptoms not assessed or unknown if assessed
Return to Schema ID Table

425 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00790: Lymphoma (excluding CLL/SLL) (2018+)
3859: HIV Status
Item Length: 1
NAACCR Item #: 3859
XML Parent-NAACCR ID: Tumor-hivStatus
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00790: Lymphoma (excluding CLL/SLL) (2018+)
00795: Lymphoma-CLL/SLL (2018+)
Description
HIV status refers to infection with the Human Immunodeficiency Virus which causes Acquired Immune
Deficiency Syndrome (AIDS). AIDS is associated with increased risk of developing some lymphomas.
Rationale
HIV status can be collected by the surveillance community for neoplasms (e.g., Kaposi Sarcoma,
Lymphomas) that are closely related to HIV/AIDS. Prior to 2018, Lymphoma SSF#1 was used for HIV
Status.
Definition
Human immunodeficiency virus (HIV) is the causal agent for acquired immune deficiency syndrome
(AIDS). Certain types of cancer are associated with HIV and AIDS, including Hodgkin lymphoma, diffuse
large B-cell lymphoma, and primary central nervous system lymphoma. These diseases in patients with
HIV or AIDS have different clinical and pathological features from the same diseases when they occur in
the general population, such as more extranodal involvement. This data item documents whether the
patient has HIV infection or AIDS at the time of diagnosis.
Coding guidelines
Code whether the patient has HIV or AIDS, based on statements in the medical record. Do not assume
that the patient is negative for HIV or AIDS unless there is a statement to that effect; code 9 instead.
Code 0 when there is a statement in the record that
o HIV or AIDS is not present
o the patient has been tested and is negative for HIV or AIDS
o the patient has been tested and is not infected with HIV or AIDS
o the malignancy is not associated with human immunodeficiency virus (HIV) or
autoimmune deficiency syndrome (AIDS)
o an HIV or AIDS test has been done and is negative
Code 1 when there is a statement in the record that
o HIV or AIDS is present
o the patient is positive for HIV or AIDS
o the patient is infected with HIV or AIDS
o the patient has a history of HIV or AIDS

426 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
o an HIV or AIDS test has been done and is positive
Code 9 when there is no mention of HIV or AIDS status in the medical record.
Additional Information
Source documents: clinical laboratory test, statement in medical record
Other names: HIV type 1, HIV type 2, ARC (AIDS related complex), PWA (person with AIDS),
PWARC (person with ARC); older terms for HIV type 1: HTLV-3, LAV
Coding Instructions and Codes
Note 1: Physician statement of HIV status can be used to code this data item when no other information
is available.
Note 2: Acquired Immune Deficiency Syndrome (AIDS) lymphomas are a late manifestation of Human
Immunodeficiency Virus (HIV) infection and have unique clinical and pathological features that differ
from lymphomas in the general population. They have a preponderance for extranodal involvement,
with central nervous system being the most common site.
Note 3: HIV includes types I and II. Older terminology includes Human T Lymphotropic Virus -3 (HTLV-3)
and Lymphadenopathy Associated Virus (LAV).
Note 4: Code 9 if there is no mention of HIV/AIDS in the medical record. Do not assume that the patient
is HIV negative.
Note 5: If patient has a history of HIV, assign code 1 even if HIV is not currently detectable.
Code Description
0 Not associated with Human Immunodeficiency Virus (HIV)/Acquired Immune Deficiency
Syndrome (AIDS)
HIV negative
1 Associated with HIV/AIDS
HIV positive
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
HIV status not assessed or unknown if assessed
Return to Schema ID Table

427 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00790: Lymphoma (excluding CLL/SLL) (2018+)
3896: NCCN International Prognostic Index (IPI)
Item Length: 2
NAACCR Item #: 3896
XML Parent-NAACCR ID: Tumor-nccnInternationalPrognosticIndex
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00790: Lymphoma (excluding CLL/SLL) (2018+)
00795: Lymphoma-CLL/SLL (2018+)
Description
The NCCN International Prognostic Index (IPI) (previously only “IPI”) is used to define risk groups for
specific lymphomas using a 0-8 score range, based on age, stage, number of extranodal sites of
involvement, patient’s performance status and LDH level.
Rationale
NCCN International Prognostic Index (IPI) is a Registry Data Collection Variable in AJCC. It was previously
collected for Lymphomas, SSF #3.
Definition
The NCCN International Prognostic Index (IPI) has been developed for lymphomas and predicts outcome
based on the following adverse factors:
Age greater than or equal to 60 years
Serum LDH greater than normal
Performance status 2-4
Stage III or IV
Extranodal involvement greater than 1 site
Additional Information
Source documents: patient history, progress notes, consultant notes, other statements in
medical record
Coding Instructions and Codes
Note 1: Physician statement of NCCN IPI must be used to code this data item. Do not calculate points or
assign risk. Only record points or risk if a physician has documented them. Use points over risk if both
are available.
Note 2: NCCN is applicable for non-Hodgkin lymphomas only.
If you have a score for Hodgkin lymphomas (IPS), do not record that information here. Code X9.
Note 3: A low, intermediate or high risk associated with Rai Stage is not recorded in this data item.

428 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description
00-08 0-8 points
X1 Stated as low risk (0-1 point)
X2 Stated as low intermediate risk (2-3 points)
X3 Stated as intermediate risk (4-5 points)
X4 Stated as high risk (6-8 points)
X8
Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code X8 will result in an edit error.)
X9
Not documented in medical record
NCCN International Prognostic Index (IPI) status not assessed or unknown if assessed
Return to Schema ID Table

430 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
Rai Classification
Definition
The Rai classification system is now used to stage chronic lymphocytic leukemia/small lymphocytic
lymphoma (CLL/SLL) (9823/3) when bone marrow and/or peripheral blood are involved using several
different criteria. The stages are based on the absence or presence of the following criteria
3885: Lymphocytosis
3804: Adenopathy
3907: Organomegaly
3811: Anemia
3933: Thrombocytopenia
Note: All of these data items are required for Staging for the AJCC Staging System Hodgkin and Non-
Hodgkin Lymphomas (9823/3 only) and EOD.
Rai stages
Stage 0 CLL is characterized by absolute lymphocytosis (>15,000/mm3) without adenopathy,
hepatosplenomegaly, anemia, or thrombocytopenia
Stage I CLL is characterized by absolute lymphocytosis with lymphadenopathy without
hepatosplenomegaly, anemia, or thrombocytopenia
Stage II CLL is characterized by absolute lymphocytosis with either hepatomegaly or
splenomegaly with or without lymphadenopathy
Stage III CLL is characterized by absolute lymphocytosis and anemia (hemoglobin <11 g/dL) with
or without lymphadenopathy, hepatomegaly, or splenomegaly
Stage IV CLL is characterized by absolute lymphocytosis and thrombocytopenia (<100,000/mm3)
with or without lymphadenopathy, hepatomegaly, splenomegaly, or anemia
Per confirmation from medical oncologists, Rai stage is only recorded for patients who have bone
marrow and/or peripheral blood involvement. Per the Hematopoietic Rules, primary site would be C421
(See Hematopoietic Manual, Module 3: Rules PH 5, 6). A new code has been added to the 5 SSDIs (code
5) to use when primary site is not C421.
A Derived Rai stage will be implemented in the 2022 updates. This new data item will take the
information from the 5 SSDIs (Lymphocytosis, Adenopathy, Organomegaly, Anemia, Thrombocytopenia),
where primary site is C421 and automatically derive the Rai stage
This table below can be used when the ONLY information available is the documented Rai stage from
the managing physician. (Note: The Rai stage cannot be found on a pathology report).
Rai
Stage
Definition Lymphocytosis Adenopathy Organomegaly Anemia Thrombocytopenia
0
Lymphocytosis only
Absolute Lymphocyte count > 5,000
cell/uL)
6 0 0 0 0
I
Lymphocytosis AND
Adenopathy
6 1 0 0 0

431 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Rai
Stage
Definition Lymphocytosis Adenopathy Organomegaly Anemia Thrombocytopenia
II
Lymphocytosis AND
Enlarged spleen and/or liver
(Organomegaly)
6 9 1 0 0
III
Lymphocytosis AND
Hemoglobin (Hgb) less than 11 g/dL
(Anemia)
6 9 9 6 0
IV
Lymphocytosis AND
Platelet count < 100,000 /uL
(Thrombocytopenia)
6 9 9 9 6
For cases with diagnosis date of 2018+ and already abstracted with a primary site not equal to C421
(bone marrow), the SSDIs will be updated with a new code of 5, which is “Not applicable: Primary site is
not C421.” A Rai Stage of 88 will then be derived.
For CLL/SLL cases abstracted after the updates and primary site not equal to C421 (bone marrow), code
the SSDIs to 5.
Return to Schema ID Table

432 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
3885: Lymphocytosis
Item Length: 1
NAACCR Item #: 3885
XML Parent-NAACCR ID: Tumor-lymphocytosis
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00795: Lymphoma-CLL/SLL (2018+)
Description
Lymphocytosis s defined by an excess of lymphocytes in the blood. In staging of Chronic Lymphocytic
Leukemia/Small Lymphocytic Leukemia (CLL/SLL), lymphocytosis is defined as an absolute lymphocyte
Rationale
Lymphocytosis is a prognostic factor required for staging of Chronic Lymphocytic Leukemia/Small
Lymphocytic Leukemia (CLL/SLL) in AJCC 8
th
edition, Chapter 79 Hodgkin and Non-Hodgkin Lymphomas.
It is a new data item for cases diagnosed 1/1/2018+.
See Rai Classification for additional information.
Coding Instructions and Codes
Note 1: For cases diagnosed 1/1/2018 and later, all cases of CLL and SLL will require both the Lugano
classification, which is captured in the AJCC stage group data item, and the five components of the
modified Rai staging system, which are captured in Site-Specific Data Items (adenopathy, anemia,
lymphocytosis, organomegaly, and thrombocytopenia)
The terms B-cell lymphocytic leukemia/chronic lymphocytic leukemia (CLL) and small lymphocytic
lymphoma (SLL) are different clinical presentations of the same disease, with both terms coded
9823. Traditionally the lymphoma diagnosis was staged with the Ann Arbor staging system and it is now
staged with the Lugano classification. In North America, CLL was staged with the Rai system.
Note 2: Rai stage is only applicable for CLL, in which the bone marrow and/or peripheral blood are
involved (primary site C421 for bone marrow, see Hematopoietic Manual, Module 3: PH 5, 6).
If primary site is not C421, code 5
Note 3: Lymphocytosis (lymphocyte number) is defined by an absolute lymphocyte count (ALC) > 5,000
Use the cut points listed in the table regardless of the lab’s reference range
For cases that document lymphocyte count in SI (Systeme Internationale) units as any of 10*9/L,
10^9/L, or 10E9/L, the cut point of 5,000 cells/µL is equivalent to (5 cells x 10*9/L), (5 cells X
10^9/L), or (5 cells x10E9/L)

433 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: Record this data item based on a blood test (CBC and differential) performed at diagnosis (pre-
treatment). In the absence of the lab test, a physician’s statement can be used.
Note 5: If there is no mention of lymphocytosis, or relevant lab results, code 9.
Note 6: The physician’s stated Rai stage always takes priority when there is conflicting information.
Code Description
0 Lymphocytosis not present
1 Lymphocytosis present
Absolute lymphocyte count > 5,000
5 Not applicable: Primary site is not C421
6 Lab value unknown, physician states lymphocytosis is present
Physician states Rai stage 0-IV
7 Test ordered, results not in chart
9 Not documented in medical record
Lymphocytosis not assessed or unknown if assessed
No Rai stage is documented in the record and there is no documentation of
lymphocytosis
Return to Schema ID Table

434 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
3804: Adenopathy
Item Length: 1
NAACCR Item #: 3804
XML Parent-NAACCR ID: Tumor-adenopathy
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00795: Lymphoma-CLL/SLL (2018+)
Description
Adenopathy is defined as the presence of lymph nodes > 1.5 cm on physical examination (PE) and is part
of the staging criteria for Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma (CLL/SLL).
Rationale
Adenopathy is a prognostic factor required for staging of CLL/SLL in AJCC 8
th
edition, Chapter 79 Hodgkin
and Non-Hodgkin Lymphomas. It is a new data item for cases diagnosed 1/1/2018+.
See Rai Classification for additional information.
Coding Instructions and Codes
Note 1: For cases diagnosed 1/1/2018 and later, all cases of CLL and SLL will require both the Lugano
classification, which is captured in the AJCC stage group data item, and the five components of the
modified Rai staging system, which are captured in Site-Specific Data Items (adenopathy, anemia,
lymphocytosis, organomegaly, and thrombocytopenia)
The terms B-cell lymphocytic leukemia/chronic lymphocytic leukemia (CLL) and small lymphocytic
lymphoma (SLL) are different clinical presentations of the same disease, with both terms coded
9823. Traditionally the lymphoma diagnosis was staged with the Ann Arbor staging system and it is now
staged with the Lugano classification. In North America, CLL was staged with the Rai system.
Note 2: Rai stage is only applicable for CLL, in which the bone marrow and/or peripheral blood are
involved (primary site C421 for bone marrow, see Hematopoietic Manual, Module 3: PH 5, 6).
If primary site is not C421, code 5
Note 3: Physician statement of presence or absence of adenopathy should be used to code this data
item.
Physician’s statement regarding the presence of adenopathy (present or absent) takes priority.
If a physician’s statement and imaging are both available and in disagreement, go with the
physician’s statement
If a physician’s statement is not available, use the definition of adenopathy in Note 3 to
determine if adenopathy is present or not

435 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: Adenopathy is defined as the presence of lymph nodes >1.5 cm on physical examination (PE)
and is part of the staging criteria.
Note 5: This data item is determined from physical exam alone. If a physical exam cannot be used to
detect adenopathy due to issues related to the patient’s obesity, a physician statement of peripheral
adenopathy based on a CT scan can be used.
A finding of retroperitoneal or mesenteric adenopathy on CT is not used in determining
adenopathy and does not affect the assigned stage
Note 6: If there is no mention of adenopathy (present or absent), code 9.
Note 7: The physician’s stated Rai stage always takes priority when there is conflicting information.
Code Description
0 Adenopathy not identified/not present
No lymph nodes > 1.5 cm
Physician states Rai Stage 0
1 Adenopathy present
Presence of lymph nodes > 1.5 cm
Physician stated Rai Stage I
5 Not applicable: Primary site is not C421
9 Not documented in medical record
Adenopathy not assessed or unknown if assessed
No Rai stage is documented in the record and there is no documentation
of adenopathy
Physician states Rai stage II-IV and there is no documentation of
adenopathy
Return to Schema ID Table

436 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
3907: Organomegaly
Item Length: 1
NAACCR Item #: 3907
XML Parent-NAACCR ID: Tumor-organomegaly
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00795: Lymphoma-CLL/SLL (2018+)
Description
Organomegaly is defined as presence of enlarged liver and/or spleen on physical examination and is part
of the staging criteria for Chronic Lymphocytic Leukemia/Small Lymphocytic Leukemia (CLL/SLL).
Rationale
Organomegaly is a prognostic factor required for staging of CLL/SLL in AJCC 8
th
edition, Chapter 79
Hodgkin and Non-Hodgkin Lymphomas. It is a new data item for cases diagnosed 1/1/2018+.
See Rai Classification for additional information.
Coding Instructions and Codes
Note 1: For cases diagnosed 1/1/2018 and later, all cases of CLL and SLL will require both the Lugano
classification, which is captured in the AJCC stage group data item, and the five components of the
modified Rai staging system, which are captured in Site-Specific Data Items (adenopathy, anemia,
lymphocytosis, organomegaly, and thrombocytopenia)
The terms B-cell lymphocytic leukemia/chronic lymphocytic leukemia (CLL) and small lymphocytic
lymphoma (SLL) are different clinical presentations of the same disease, with both terms coded
9823. Traditionally the lymphoma diagnosis was staged with the Ann Arbor staging system and it is now
staged with the Lugano classification. In North America, CLL was staged with the Rai system.
Note 2: Rai stage is only applicable for CLL, in which the bone marrow and/or peripheral blood are
involved (primary site C421 for bone marrow, see Hematopoietic Manual, Module 3: PH 5, 6).
If primary site is not C421, code 5
Note 3: Physician statement of presence or absence of organomegaly should be used to code this data
item.
Note 4: Organomegaly is defined as presence of enlarged liver and/or spleen on physical examination
and is part of the staging criteria.
Note 5: This data item is determined from physical exam alone. If a physical exam cannot be used to
detect organomegaly due to issues related to the patient’s obesity, a physician statement of
organomegaly based on a CT scan can be used.

437 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 6: I If there is no mention of the presence or absence of organomegaly (hepatomegaly and
splenomegaly), code 9.
Both the liver and spleen mut be evaluated and determined to be normal to code 0. If only one
is evaluated and determined to be normal, code 9.
Note 7: The physician’s stated Rai stage always takes priority when there is conflicting information.
Code Description
0 Neither hepatomegaly (liver) nor splenomegaly (spleen) present
Physician states Rai Stage 0-I
1 Hepatomegaly (liver) and/or splenomegaly (spleen) present
Physician states Rai Stage II
5 Not applicable: Primary site is not C421
9 Not documented in medical record
Organomegaly (hepatomegaly and/or splenomegaly) not assessed or
unknown if assessed
No Rai stage is documented in the record and there is no documentation
of organomegaly
Physician states Rai stage III-IV and there is no documentation of
organomegaly
Return to Schema ID Table

438 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
3811: Anemia
Item Length: 1
NAACCR Item #: 3811
XML Parent-NAACCR ID: Tumor-anemia
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00795: Lymphoma-CLL/SLL (2018+)
Description
Anemia is defined by a deficiency of red blood cells or of hemoglobin in the blood. In staging of Chronic
Lymphocytic Leukemia/Small Lymphocytic Leukemia (CLL/SLL), anemia is defined as Hgb <11.0 g/dL.
Rationale
Anemia is a prognostic factor required for staging of Chronic Lymphocytic Leukemia/Small Lymphocytic
Leukemia (CLL/SLL) in AJCC 8
th
edition, Chapter 79 Hodgkin and Non-Hodgkin Lymphomas. It is a new
data item for cases diagnosed 1/1/2018+.
See Rai Classification for additional information.
Coding Instructions and Codes
Note 1: For cases diagnosed 1/1/2018 and later, all cases of CLL and SLL will require both the Lugano
classification, which is captured in the AJCC stage group data item, and the five components of the
modified Rai staging system, which are captured in Site-Specific Data Items (adenopathy, anemia,
lymphocytosis, organomegaly, and thrombocytopenia)
The terms B-cell lymphocytic leukemia/chronic lymphocytic leukemia (CLL) and small lymphocytic
lymphoma (SLL) are different clinical presentations of the same disease, with both terms coded
9823. Traditionally the lymphoma diagnosis was staged with the Ann Arbor staging system and it is now
staged with the Lugano classification. In North America, CLL was staged with the Rai system.
Note 2: Rai stage is only applicable for CLL, in which the bone marrow and/or peripheral blood are
involved (primary site C421 for bone marrow, see Hematopoietic Manual, Module 3: PH 5, 6).
If primary site is not C421, code 5
Note 3: Anemia is defined as Hgb <11.0 g/dL and is part of the staging criteria.
Use the cut points listed in the table regardless of the lab’s reference range
A lab value expressed in grams per liter (g/L) is 10 times the same value expressed in g/dL;
therefore, the cut point of 11.0 g/dL is equivalent to 110 g/L
Note 4: Record this data item based on a blood test (CBC, hemoglobin & hematocrit, H&H) performed at
diagnosis (pre-treatment). In the absence of the lab test, a physician’s statement can be used.
Note 5: If there is no mention of anemia, or relevant lab results, code 9.

439 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 6: The physician’s stated Rai Stage always takes priority when there is conflicting information.
Code Description
0 Anemia not present
Hgb >=11.0 g/dL
Physician states Rai Stage 0-II
1 Anemia present
Hgb <11.0 g/dL
5 Not applicable: Primary site is not C421
6 Lab value unknown, physician states patient is anemic
Physician states Rai Stage III
7 Test ordered, results not in chart
9 Not documented in medical record
Anemia not assessed or unknown if assessed
No Rai stage is documented in the record and there is no
documentation of anemia
Physician states Rai stage IV and there is no documentation of anemia
Return to Schema ID Table

440 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
3933: Thrombocytopenia
Item Length: 1
NAACCR Item #: 3933
XML Parent-NAACCR ID: Tumor-thrombocytopenia
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00795: Lymphoma-CLL/SLL (2018+)
Description
Thrombocytopenia is defined by a deficiency of platelets in the blood. In staging of Chronic Lymphocytic
Leukemia/Small Lymphocytic Leukemia (CLL/SLL), thrombocytopenia is defined as Platelets (Plt) <
100,000/ L.
Rationale
Thrombocytopenia is a prognostic factor required for staging of Chronic Lymphocytic Leukemia/Small
Lymphocytic Leukemia (CLL/SLL) in AJCC 8
th
edition, Chapter 79 Hodgkin and Non-Hodgkin Lymphomas.
It is a new data item for cases diagnosed 1/1/2018+.
See Rai Classification for additional information.
Coding Instructions and Codes
Note 1: For cases diagnosed 1/1/2018 and later, all cases of CLL and SLL will require both the Lugano
classification, which is captured in the AJCC stage group data item, and the five components of the
modified Rai staging system, which are captured in Site-Specific Data Items (adenopathy, anemia,
lymphocytosis, organomegaly, and thrombocytopenia)
The terms B-cell lymphocytic leukemia/chronic lymphocytic leukemia (CLL) and small lymphocytic
lymphoma (SLL) are different clinical presentations of the same disease, with both terms coded
9823. Traditionally the lymphoma diagnosis was staged with the Ann Arbor staging system and it is now
staged with the Lugano classification. In North America, CLL was staged with the Rai system.
Note 2: Rai stage is only applicable for CLL, in which the bone marrow and/or peripheral blood are
involved (primary site C421 for bone marrow, see Hematopoietic Manual, Module 3: PH 5, 6).
If primary site is not C421, code 5
Note 3: Thrombocytopenia is defined as platelets (Plt) <100,000/ L. This is part of the Modified Rai
Staging System and not included as part of the AJCC Lugano staging.
Use the cut points listed in the table regardless of the lab’s reference range
For cases that document platelet count in SI (Systeme Internationale) units as any of 10*9/L,
10^9/L, or 10E9/L, the cut point of 100,000 cells/µL is equivalent to (100 cells x 10*9/L), (100
cells x 10^9/L, or (100 cells x 10E9/L)

441 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Note 4: Record this data item based on a blood test (CBC and differential) performed at diagnosis (pre-
treatment). In the absence of the lab test, a physician’s statement can be used.
Note 5: If there is no mention of thrombocytopenia, or the relevant lab tests, code 9.
Note 6: The physician’s stated Rai Stage always takes priority when there is conflicting information.
Code Description
0 Thrombocytopenia not present
Platelets (Plt) >=100,000/ L
Physician states Rai Stage 0-III
1
Thrombocytopenia present
Platelets (Plt) < 100,000/ L
5 Not applicable: Primary site is not C421
6 Lab value unknown, physician states thrombocytopenia is present
Physician states Rai Stage IV
7 Test ordered, results not in chart
9 Not documented in medical record
Thrombocytopenia not assessed or unknown if assessed
No Rai stage is documented in the record and there is no documentation
of thrombocytopenia
Return to Schema ID Table

442 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00795: Lymphoma-CLL/SLL (2018+)
3955: Derived Rai stage
Item Length: 1
NAACCR Item #: 3955
XML Parent-NAACCR ID: Tumor-derivedRaiStage
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00795: Lymphoma-CLL/SLL (2018+)
Description
This data item stores the Derived Rai stage value derived from the values coded in the following SSDIs
for the Lymphoma-CLL/SLL schema (9823/3).
3885: Lymphocytosis
3804: Adenopathy
3907: Organomegaly
3811: Anemia
3933: Thrombocytopenia
The Rai stage is only applicable for Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma
(CLL/SLL) (9823/3) cases where the primary site is bone marrow (C421). For cases with a primary site
other than bone marrow (C421), the derived Rai stage will be 8 and all the SSDIs will be coded to 5.
Derivation will be run on all cases diagnosed 1/1/2018 and forward.
Rationale
The Derived Rai stage can be used to evaluate disease spread at diagnosis, treatment patterns and
outcomes over time.
Rai Stage Rai Code
Description
0 0 Lymphocytosis
1 I Lymphocytosis and Adenopathy
2
II
Lymphocytosis and
Organomegaly
(Adenopathy is any value other than 5)
3
III
Lymphocytosis and Anemia
(Adenopathy and Organomegaly are any value other than 5)
4
IV
Lymphocytosis and Thrombocytopenia
(Adenopathy, Organomegaly and Anemia are any value other than 5)
8 N/A Does not apply, primary site not bone marrow (C421)
(All 5 SSDIs should be set to 5)
9
9
Unknown(All 5 SSDIs are 9 or blank
)
At least one is set to 9 OR
Lymphocytosis is 0,7,9 OR
Lymphocytosis is blank and one of the other SSDIs is a value other than 5 or 9)
This field should be left blank for all cases diagnosed prior to 2018, for schemas other than 00795, and
when not required by standard setter.

443 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00811: Mycosis Fungoides (2018+)
3910: Peripheral Blood Involvement
Item Length: 1
NAACCR Item #: 3910
XML Parent-NAACCR ID: Tumor-peripheralBloodInvolvement
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00811: Mycosis Fungoides (2018+)
Description
Peripheral blood involvement, summarized in “B category”, refers to the percentage of peripheral blood
lymphocytes that are atypical (Sezary) cells and whether they are “Clone negative” or “Clone positive.”
Rationale
Peripheral blood involvement is a prognostic factor required in AJCC 8
th
edition, Chapter 81 Primary
Cutaneous Lymphomas, for staging of Mycosis Fungoides and Sezary Syndrome. It was previously
collected as Mycosis Fungoides, CS SSF #1.
Definition
Mycosis fungoides is the most common type of primary cutaneous T-cell lymphoma. Sezary syndrome is
a more aggressive type of primary cutaneous T-cell lymphoma in which a specific type of malignant T
lymphocytes (Sezary cells) are present in the circulating blood. Staging of mycosis fungoides includes
analysis of the circulating blood for Sezary cells. This analysis can be done by microscopy or flow
cytometry. Results of microscopy are reported as counts of Sezary cells per cubic millimeter or the
percentage of Sezary cells as a proportion of total lymphocytes. Flow cytometry looks for specific cell
surface markers such as CD26.
Information about peripheral blood involvement and T-cell clonality identified by polymerase chain
reaction (PCR) or Southern blot analysis is combined in a “B” category unique to mycosis fungoides
staging in the TNM system.
The basic categories are B0 (no significant blood involvement); B1 (low blood tumor burden); and B2
(high blood tumor burden). Any mention of B2 puts the case into Stage IV. B0 and B1 are
subcategorized by clonality.
Code a statement of peripheral blood involvement and clonality (if given) as reported by the clinician
from tissue and/or blood samples. If the physician does not provide a B rating but counts or
percentages of neoplastic cells, flow cytometry test results, and/or clonality test results are performed,
use the appropriate code for the amount of blood involvement with “clone unknown.”
Additional Information
Source documents: pathology report, clinical laboratory reports of blood analysis (tissue and
blood samples)

444 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Other names: Peripheral blood involvement: circulating Sezary cells, T-cell clonality: T-cell
receptor (TCR) gene rearrangement, Monoclonal: clone +, clone positive, Polyclonal: clone –,
clone negative
Coding Instructions and Codes
Note 1: The categories for peripheral blood involvement (B rating) are
B0: No significant blood involvement
B1: Low blood tumor burden
B2: High blood tumor burden
Note 2: Physician statement of B rating can be used to code this data item.
Note 3: If counts or percentages of neoplastic cells and clonality test results are available, but a B rating
is not stated by the physician, the registrar can use the information and assign a B rating and code this
data item accordingly. If this information is not available, code 9.
Code Description B Map
0 Absence of significant blood involvement
5% or less of peripheral blood lymphocytes are atypical (Sezary) cells
Clone unknown
Stated as B0
B0
1 Absence of significant blood involvement
5% or less of peripheral blood lymphocytes are atypical (Sezary) cells
Clone negative
Stated as B0a
B0a
2 Absence of significant blood involvement:
5% or less of peripheral blood lymphocytes are atypical (Sezary) cells
Clone positive
Stated as B0b
B0b
3 Low blood tumor burden
More than 5% of peripheral blood lymphocytes are atypical (Sezary) cells
but does not meet the criteria of B2
Clone unknown
Stated as B1
B1
4 Low blood tumor burden
More than 5% of peripheral blood lymphocytes are atypical (Sezary) cells
but does not meet the criteria of B2
Clone negative
Stated as B1a
B1a
5 Low blood tumor burden
More than 5% of peripheral blood lymphocytes are atypical (Sezary) cells
but does not meet the criteria of B2
Clone positive
Stated as B1b
B1b

445 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code Description B Map
6 High blood tumor burden
Greater than or equal to 1000 Sezary cells per microliter (uL)
Clone positive
Stated as B2
B2
7 Test ordered, results not in chart BX
9
Not documented in
medical record
Peripheral Blood Involvement not assessed or unknown if assessed
BX
Return to Schema ID Table

446 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00821: Plasma Cell Myeloma (2018+)
RISS Stage (Plasma Cell Myeloma)
Definition
The Revised International Staging System (RISS or R-ISS) is now used to stage plasma cell myeloma
(9732/3), using several different criteria. The stages are based on the absence or presence of the
following criteria
3931: Serum Beta-2 Microglobulin Pretreatment Level
3930: Serum Albumin Pretreatment Level
3857: High Risk Cytogenetics
3869: LDH Level
Required for Staging: The AJCC Staging System Plasma Cell Myeloma and Plasma Cell Disorders (9732/3
only) and EOD.
Note: RISS stage is not applicable for the descriptions of plasma multiple myeloma that are
listed in the codes 1 and 9 in the SSDI Schema Discriminator 1: Plasma Cell Myeloma
Terminology. If you have coded 1 or 9 for this Schema Discriminator, the four data items listed
above are BLANK.
The RISS stages are
Stage I: Serum Beta-2-microglobulin <3.5 mg/L and serum albumin > 3.5 g/dL and no high-risk
cytogenetics and Normal LDH
Stage II: Not R-ISS I or III
Stage III: Serum Beta-2-microglobulin > 5.5 mg/L and high-risk cytogenetics and/or high LDH
Additional Information
Other names: R-ISS
Return to Schema ID Table

447 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00821: Plasma Cell Myeloma (2018+)
3926: Schema Discriminator 1: Plasma Cell Myeloma Terminology
Item Length: 1
NAACCR Item #: 3926
XML Parent-NAACCR ID: Tumor-schemaDiscriminator1
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00821: Plasma Cell Myeloma (2018+)
Definition
A variety of descriptive terms refer to early phases of plasma cell myeloma, all of which are coded to
9732, and reportable based on the 2010 Hematopoietic and Lymphoid Neoplasms coding rules. Per AJCC
8
th
edition, not all terms are applicable for the Revised International Staging System (RISS or R-ISS) stage.
This schema discriminators collects the specific terminology used to describe the plasma cell myeloma
at the time of diagnosis.
Code the terminology used by the physician to describe the plasma cell myeloma from any
documentation in the medical record. If other terminology is used later in the course of the disease to
describe more aggressive plasma cell myeloma, do not change the code in the schema discriminator.
Coding Instructions and Codes
Note 1: Several terms are used to characterize plasma cell myeloma at the time of diagnosis. All these
terms are reportable according to the new Hematopoietic and Lymphoid Neoplasms rules effective for
cases diagnosed January 1, 2010 and later.
Note 2: Select the code based on the terminology specified by the physician in the record. Do not
attempt to determine the correct terminology based on the diagnostic criteria in the AJCC 8
th
table 82.1.
Note 3: Do not change the discriminator code if a term used later indicates progression to a more
aggressive disease course.
Note 4: If diagnosis is plasma cell leukemia variant and is diagnosed concomitant with plasma cell
myeloma, code 0.
Code Description Stage Table
0 Plasma cell myeloma (PCM)
Multiple myeloma
Myeloma, NOS
Non-secretory myeloma
Ultra-High-Risk Smoldering MM (SMM)
RISS Stage
1 Smoldering plasma cell myeloma (SPCM)
Asymptomatic plasma cell myeloma
Early myeloma
Evolving myeloma
No RISS Stage
9 Other terminology describing myeloma
Unknown terminology used
No RISS Stage
Return to Schema ID Table

448 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00821: Plasma Cell Myeloma (2018+)
3931: Serum Beta-2 Microglobulin Pretreatment Level
Item Length: 1
NAACCR Item #: 3931
XML Parent-NAACCR ID: Tumor-serumBeta2MicroglobulinPretxLvl
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00821: Plasma Cell Myeloma (2018+)
Description
Serum Beta-2 Microglobulin is a protein that is found on the surface of many cells and plentiful on the
surface of whi -2)
-2) Microglobulin level is a prognostic factor for
plasma cell myeloma.
Rationale
Serum Beta-2 Microglobulin Pretreatment Level is a prognostic factor required in AJCC 8
th
edition,
Chapter 82 Plasma Cell Myeloma and Plasma Cell Disorders, for staging of plasma cell myeloma. It is a
new data item for cases diagnosed 1/1/2018+.
See RISS Stage (Plasma Cell Myeloma) for additional information.
Coding Instructions and Codes
Note 1: Serum microglobulin is part of the Revised International Staging (RISS). Use the cut points listed
in the table below regardless of the lab’s reference range.
Note 2: Record this data item based on a blood test performed at diagnosis (pre-treatment). In the
absence of the lab test, a physician’s statement of the exact value can be used. Use the highest value
available.
Note 3: If there is no mention of the serum beta-2 microglobulin, code 9.
Note 4: If Schema Discriminator 1: Plasma Cell Myeloma Terminology is coded to 1 or 9, code 5.
Code Description
0 -microglobulin < 3.5 mg/L
1 -
2 -
5 Schema Discriminator 1: Plasma Cell Myeloma Terminology coded to 1 or 9
7 Test ordered, results not in chart
9 Not documented in medical record
Serum Beta-2 Microglobulin Pretreatment Level not assessed or unknown
if assessed
Return to Schema ID Table

449 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00821: Plasma Cell Myeloma (2018+)
3930: Serum Albumin Pretreatment Level
Item Length: 1
NAACCR Item #: 3930
XML Parent-NAACCR ID: Tumor-serumAlbuminPretreatmentLevel
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00821: Plasma Cell Myeloma (2018+)
Description
Albumin is the most abundant protein in human blood plasma. Serum albumin pretreatment level is a
prognostic factor for plasma cell myeloma.
Rationale
Serum albumin pretreatment level is a prognostic factor required in AJCC 8
th
edition, Chapter 82 Plasma
Cell Myeloma and Plasma Cell Disorders, for the Revised International Staging System (RISS). It is a new
data item for cases diagnosed 1/1/2018+.
See RISS Stage (Plasma Cell Myeloma) for additional information.
Coding Instructions and Codes
Note 1: 3.5 g/dL and is part of the Revised International Staging
System (RISS).
Use the cut points listed in the table regardless of the lab’s reference range
A lab value expressed in grams per liter (g/L) is 10 times the same value expressed in g/dL;
therefore, the cut point of 3.5 g/dL is equivalent to 35 g/L.
Note 2: Record this data item based on a blood test performed at diagnosis (pre-treatment). In the
absence of the lab test, a physician’s statement of the exact value can be used. Do not use findings from
a urine test.
Note 3: If there is no mention of the serum albumin, code 9.
Note 4: If Schema Discriminator 1: Plasma Cell Myeloma Terminology is coded to 1 or 9, code 5.
Code Description
0 Serum albumin <3.5 g/dL
1
5 Schema Discriminator 1: Plasma Cell Myeloma Terminology coded to 1 or 9
7 Test ordered, results not in chart
9 Not documented in medical record
Serum Albumin Pretreatment Level not assessed or unknown if assessed
Return to Schema ID Table

450 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00821: Plasma Cell Myeloma (2018+)
3857: High Risk Cytogenetics
Item Length: 1
NAACCR Item #: 3857
XML Parent-NAACCR ID: Tumor-highRiskCytogenetics
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00821: Plasma Cell Myeloma (2018+)
Description
High Risk Cytogenetics is defined as one or more of t(4;14), t(14;16), or del 17p identified from FISH test
results and is part of the staging criteria for plasma cell myeloma.
Rationale
High Risk Cytogenetics is a prognostic factor required in AJCC 8
th
edition, Chapter 82 Plasma Cell
Myeloma and Plasma Cell Disorders, for staging of plasma cell myeloma. It is a new data item for cases
diagnosed 1/1/2018+.
See RISS Stage (Plasma Cell Myeloma) for additional information.
Coding Instructions and Codes
Note 1: Physician statement of presence or absence of high-risk cytogenetics can be used to code this
data item.
Note 2: Record this data item based on physician statement or FISH test interpretation performed at
diagnosis (pre-treatment)
Note 3: If the presence/absence of high-risk cytogenetics determined by available test results differs
from the physician statement of presence/absence, the physician’s statement takes precedence.
Note 4: If there is no mention of high risk cytogenetics, code 9.
Note 5: If Schema Discriminator 1: Plasma Cell Myeloma Terminology is coded to 1 or 9, code 5.
Code Description
0 High-risk cytogenetics not identified/not present
1 High-risk cytogenetics present
5 Schema Discriminator 1: Plasma Cell Myeloma Terminology coded to 1 or 9
7 Test ordered, results not in chart
9 Not documented in medical record
High Risk Cytogenetics not assessed or unknown if assessed
Return to Schema ID Table

451 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00821: Plasma Cell Myeloma (2018+)
3869: LDH Level
Item Length: 1
NAACCR Item #: 3869
XML Parent-NAACCR ID: Tumor-ldhPretreatmentLevel
NAACCR Alternate Name: LDH (Lactate Dehydrogenase) Pretreatment Level
Active years: 2018+
Schema(s):
00821: Plasma Cell Myeloma (2018+)
Description
LDH (Lactate Dehydrogenase) is an enzyme involved in conversion of sugars to energy and present in
most cells in the body. Elevated LDH is an adverse prognostic factor for plasma cell myeloma and
melanoma of the skin.
Rationale
LDH (Lactate Dehydrogenase) Level is a prognostic factor required in AJCC 8
th
edition for Chapter 82
Plasma Cell Myeloma and Plasma Cell Disorders and Chapter 47 Melanoma Skin. For Plasma Cell
Myeloma, LDH is part of the RISS Stage and is new for cases diagnosed 1/1/2018+. For Melanoma Skin,
LDH is used to define the M subcategories and was previously collected as Melanoma Skin, SSF #4.
See RISS Stage (Plasma Cell Myeloma) for additional information.
Coding Instructions and Codes
Note 1: Use the reference ranges from your lab to determine if LDH is normal.
Note 2: Record this data item based on a blood test performed at diagnosis (pre-treatment). In the
absence of the lab test, a physician’s statement of the exact value or interpretation can be used. Use the
highest value available.
Note 3: If there is no mention of the LDH, code 9.
Note 4: If Schema Discriminator 1: Plasma Cell Myeloma Terminology is coded to 1 or 9, code 5.
Code Description
0 Normal LDH level
Low, below normal
1 Above normal LDH level; High
5 Schema Discriminator 1: Plasma Cell Myeloma Terminology coded to 1 or 9
7 Test ordered, results not in chart
9 Not documented in medical record
LDH (Lactate Dehydrogenase) Level not assessed or unknown if assessed
Return to Schema ID Table

453 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
00830: HemeRetic (2018+)
3862: JAK 2
Item Length: 1
NAACCR Item #: 3862
XML Parent-NAACCR ID: Tumor-jak2
NAACCR Alternate Name: None
Active years: 2018+
Schema(s):
00830: HemeRetic (2018+)
Description
Janus Kinase 2 (JAK2, JAK 2) is a gene mutation that increases susceptibility to several myeloproliferative
neoplasms (MPNs). Testing for the JAK2 mutation is done on whole blood. Nearly all people with
polycythemia vera, and about half of those with primary myelofibrosis and essential thrombocythemia,
have the mutation. JAK2 analysis continues to increase in use for hematopoietic neoplasms.
Rationale
JAK2 can be collected by the surveillance community for myeloproliferative neoplasms. Prior to 2018,
HemeRetic SSF#1 was used for JAK2.
Definition
JAK2, a gene found in all humans, is involved in the development of blood cells. If JAK2 has mutated, the
person is more susceptible to develop a myeloproliferative disorder (MPD). The JAK2 mutation, which is
acquired rather than inherited, is found in as many as 90% of patients with polycythemia vera (PV),
about half of patients with essential thrombocythemia (ET), and slightly fewer patients with primary
myelofibrosis (also known as agnogenic myeloid metaplasia and other terms). JAK2 is used by clinicians
to help classify MPDs. The most common histologies for which JAK-2 is tested are those listed above.
Registrars can use JAK2 information to help determine whether the MPD is reportable. JAK2 positivity
indicates a malignant (clonal, irreversible) reportable disease, but is not diagnostic of a specific MPD.
Additional tests, such as a bone marrow biopsy, are necessary to determine the specific MPD histology.
As the use of JAK2 increases and is investigated for other hematopoietic histologies, it also has future
potential for development of targeted therapeutics for the MPDs.
The principal JAK2 test looks for a change (mutation) in an amino acid at a specific place on the JAK2
gene called V617F. If the V617F test is negative, other JAK2 mutation tests, such as those in exon 12 or
13 may be ordered to investigate a possible diagnosis of polycythemia vera. (An exon is a segment of a
gene that contains instructions for making a protein.)
Coding guidelines
Code the result of the JAK2 test as documented in a laboratory test or elsewhere in the medical record.
Code this field for any hematopoietic, immunoproliferative, myeloproliferative, or myelodysplastic
disease for which JAK2 is tested. For those diseases where JAK2 is not mentioned in the record, or for a
HemeRetic schema disease such as leukemia where JAK2 is not normally tested, code as 9.

454 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Code 0 when the JAK2 test result is stated as negative.
Code 1 when the JAK2 test was performed and was positive for mutation V617F in exon 14.
Code 2 when the JAK2 test was performed and was positive for mutation of exon 12.
Code 3 when the JAK2 test was performed and was positive for another specified mutation.
Code 4 when the JAK2 test was performed and was positive for more than one mutation.
Code 7 when there is a statement in the record that the test was ordered but the results are not
available.
Code 9 when
o There is no information in the medical record about JAK2 testing
o The results of JAK2 testing are unknown
Additional Information
Source documents: clinical laboratory test (whole blood), reference laboratory test; anatomic
pathology (polymerase chain reaction test on bone marrow)
Other names: Janus kinase 2 gene, JAK2 V617F, JAK2 exon 12, JAK2 exon13
Coding Instructions and Codes
Note 1: Physician statement of JAK2 can be used to code this data item when no other information is
available.
Note 2: Janus Kinase 2 (JAK2, JAK 2) is a gene mutation that increases susceptibility to several
myeloproliferative neoplasms (MPNs). Testing for the JAK2 mutation is done on whole blood. Nearly all
people with polycythemia vera, and about half of those with primary myelofibrosis and essential
thrombocythemia, have the mutation.
Note 3: Record JAK2 for any hematopoietic neoplasm. It is most commonly used for the following
histologies:
Polycythemia Vera (9950/3)
Primary myelofibrosis (9961/3)
Essential Thrombocytopenia (9962/3)
Chronic myelomonocytic leukemia (9945/3)
Code Description
0 JAK2 result stated as negative
1 JAK2 positive for mutation V617F WITH or WITHOUT other mutations
2 JAK2 positive for exon 12 mutation
3 JAK2 positive for other specified mutation
4 JAK2 positive for more than one mutation other than V617F
5
JAK2 positive NOS
Specific mutation(s) not stated
7 Test ordered, results not in chart
8 Not applicable: Information not collected for this case
(If this item is required by your standard setter, use of code 8 will result in an edit error.)
9 Not documented in medical record
JAK2 not assessed or unknown if assessed
Return to Schema ID Table
456 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
ALPHABETICAL INDEX
Adenoid Cystic Basaloid Pattern, 394
Adenopathy (Rai Classification: CLL/SLL), 427
AFP Post-Orchiectomy Lab Value, 338
AFP Post-Orchiectomy Range, 340
AFP Pre-Orchiectomy Lab Value, 334
AFP Pre-Orchiectomy Range, 336
AFP Pretreatment Interpretation, 120
AFP Pretreatment Lab Value, 119
Anemia (Rai Classification: CLL/SLL), 431
B Symptoms, 416
Bilirubin Pretreatment Total Lab Value, 124
Bilirubin Pretreatment Unit of Measure, 126
Bone Invasion, 164
Brain Molecular Markers, 398
Breslow Tumor Thickness, 185
CA-125 Pretreatment Interpretation, 298
CEA Pretreatment Interpretation, 96
CEA Pretreatment Lab Value, 94
Chromosome 19q: Loss of Heterozygosity (LOH),
403
Chromosome 1p: Loss of Heterozygosity (LOH),
401
Chromosome 3 Status, 375
Chromosome 8q Status, 377
Circumferential Resection Margin (CRM), 98
Creatinine Pretreatment Lab Value, 127
Creatinine Pretreatment Unit of Measure, 129
Esophagus and EGJ Tumor Epicenter, 86
Estrogen Receptor Percent Positive or Range,
204
Estrogen Receptor Summary, 202
Estrogen Receptor Total Allred Score, 206
Extranodal Extension Clin (non-Head and Neck),
177
Extranodal Extension Head and Neck Clinical, 56
Extranodal Extension Path (non-Head and
Neck), 179
Extravascular Matrix Patterns, 379
Fibrosis Score, 131
FIGO Stage (Adenosarcoma), 295
FIGO: Cervix, 274
FIGO: Corpus Carcinoma and Carcinosarcoma,
279
FIGO: Gestational Trophoblastic Tumors
(Placenta), 304
FIGO: Ovary, Fallopian Tube, and Peritoneal
Carcinoma, 296
FIGO: Vagina, 266
FIGO: Vulva, 254
Gestational Trophoblastic Prognostic Scoring
Index, 305
Gleason Patterns Clinical, 315
Gleason Patterns Pathological, 320
Gleason Score Clinical, 318
Gleason Score Pathological, 323
Gleason Tertiary Pattern, 325
hCG Post-Orchiectomy Lab Value, 346
hCG Post-Orchiectomy Range, 348
hCG Pre-Orchiectomy Lab Value, 344
hCG Pre-Orchiectomy Range, 345
HER2 IHC Summary, 217
HER2 ISH Dual Probe Copy Number, 225
HER2 ISH Dual Probe Ratio, 227
HER2 ISH Single Probe Copy Number, 223
HER2 ISH Summary, 219
HER2 Overall Summary, 221
Heritable Trait, 390
High Risk Cytogenetics, 443
High Risk Histologic Features, 79
HIV Status, 418
International Normalized Ratio, 130
Invasion Beyond Capsule, 360
Ipsilateral Adrenal Gland Involvement, 364
JAK 2, 446
Ki-67, 242
KIT Gene Immunohistochemistry, 173
KRAS, 101
LDH Post-Orchiectomy Range, 353
LDH Pre-Orchiectomy Range, 352
LDH Pretreatment Lab Value, 192
LDH Pretreatment Level, 194
LDH Upper Limits of Normal, 195
LN Assessment Method Femoral-Inguinal, 257
LN Assessment Method Para-Aortic, 267, 269
LN Assessment Method Pelvic, 261
LN Distant Assessment Method, 272
LN Distant: Mediastinal, Scalene, 271
LN Head and Neck Levels I-III, 61
LN Head and Neck Levels IV-V, 63
LN Head and Neck Levels VI-VII, 65
LN Head and Neck Other, 67
LN Isolated Tumor Cells (ITC), 181
LN Laterality, 263
LN Positive Axillary Level I-II, 244
LN Status Femoral-Inguinal, Para-Aortic, Pelvic,
255, 259
Lymphocytosis (Rai Classification: CLL/SLL), 425
Major Vein Involvement, 362
457 | P a g e Site-Specific Data Item (SSDI) Manual Version 3.0
Measured Basal Diameter, 381
Measured Thickness, 383
Methylation of O6-Methylguanine-
Methyltransferase, 405
Microsatellite Instability (MSI), 104
Microvascular Density, 385
Mitotic Count Uveal Melanoma, 387
Mitotic Rate Melanoma, 190
Multigene Signature Method, 231
Multigene Signature Results, 233
NCCN International Prognostic Index (IPI), 420
Number of Cores Examined, 329
Number of Cores Positive, 327
Number of Examined Para-Aortic Nodes, 284
Number of Examined Pelvic Nodes, 288
Number of Positive Para-Aortic Nodes, 282
Number of Positive Pelvic Nodes, 286
Oncotype Dx Recurrence Score-DCIS, 237
Oncotype Dx Recurrence Score-Invasive, 239
Oncotype Dx Risk Level-DCIS, 238
Oncotype Dx Risk Level-Invasive, 241
Organomegaly (Rai Classification: CLL/SLL), 429
Percent Necrosis Post Neoadjuvant, 160
Perineural Invasion, 107
Peripheral Blood Involvement, 436
Peritoneal Cytology, 290
Pleural Effusion, 157
Primary Sclerosing Cholangitis, 134
Profound Immune Suppression, 183
Progesterone Receptor Percent Positive or
Range, 210
Progesterone Receptor Summary, 208
Progesterone Receptor Total Allred Score, 212
PSA (Prostatic Specific Antigen) Lab Value, 309
Residual Tumor Volume Post Cytoreduction,
300
Response to Neoadjuvant Therapy, 247
S Category Clinical, 355
S Category Pathological, 357
Sarcomatoid Features, 366
Schema Discriminator 1 (Histology
Discriminator for 9591/3), 415
Schema Discriminator 1:
BileDuctsDistal/BileDuctsPerihilar/CysticDuct,
138
Schema Discriminator 1: Lacrimal Gland/Sac,
393
Schema Discriminator 1: Melanoma Ciliary
Body/Melanoma Iris, 374
Schema Discriminator 1:
Nasopharynx/Pharyngeal Tonsil, 73
Schema Discriminator 1: Occult Head and Neck
Lymph Nodes, 49
Schema Discriminator 1: Plasma Cell Myeloma
Terminology, 440
Schema Discriminator 1: Primary Peritoneum
Tumor, 172
Schema Discriminator 1: Thyroid
Gland/Thyroglossal Duct, 412
Schema Discriminator 1: Urethra/Prostatic
Urethra, 368
Schema Discriminator 2: Histology Discriminator
for 8020/3, 85
Schema Discriminator 2: Oropharyngeal p16, 75
Separate Tumor Nodules, 147
Serum Albumin Pretreatment Level, 442
Serum Beta-2 Microglobulin Pretreatment
Level, 441
Thrombocytopenia (Rai Classification: CLL/SLL),
433, 435
Tumor Deposits, 109
Tumor Growth Pattern, 136
Ulceration, 188
Visceral and Parietal Pleural Invasion, 150, 153


















